Table of Contents
About
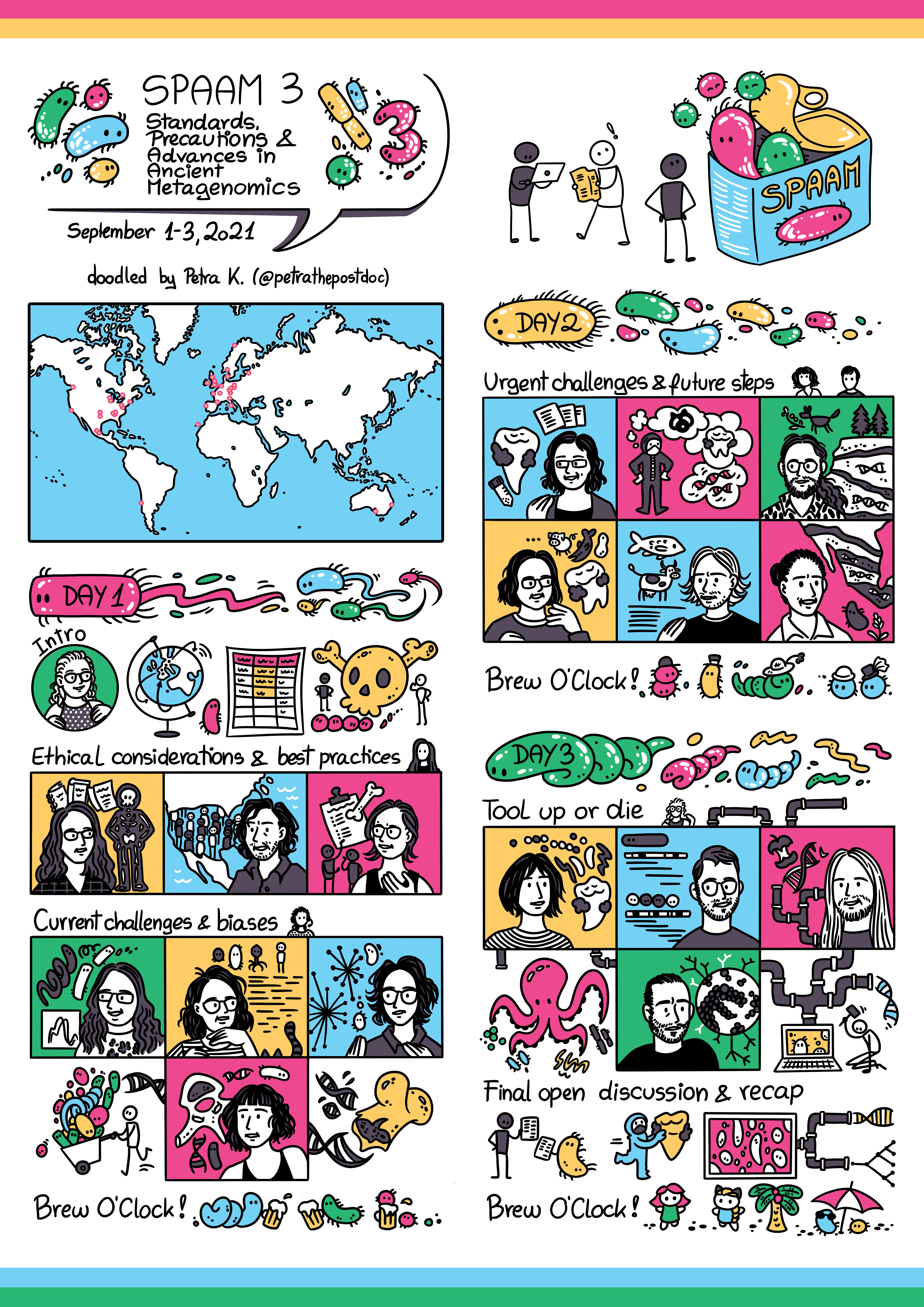
The third installment of the Standards, Precautions, and Advances in Ancient Metagenomics (SPAAM3) took place on the 1st-3rd of September 2021.
What
The third installment of the Standards, Precautions, and Advances in Ancient Metagenomics (SPAAM), aka SPAAM3, which specialises in tackling the challenges of ancient metagenomics
⚠️ SPAAM3 Registration is now CLOSED.
⚠️ the programme is now up! Please see for more information.
When
Dates
Event: 1st-3rd September 2021
Times
Starting at 6:00 PT/15:00 CET
Who
A list of participants will be made available via email.
Where
Online via Zoom video conference. Links to the event will be sent out via email!
Event Description
We are excited to announce the third instalment of the Standards, Precautions, and Advances in Ancient Metagenomics (SPAAM). The last SPAAM meeting took place virtually in September 2020, which brought together many members of the ancient microbial genomics community to encourage collaboration and knowledge sharing to tackle some of the most pressing challenges in the field. We are continuing its legacy and hope that SPAAM3 will bring researchers together for the development of effective solutions to address current methodological issues in ancient metagenomics and push the field forward. Like its predecessor, this edition will provide a platform for early career scholars to discuss their experiences and perspectives related to ancient metagenomic research. It will be held virtually on September 1st-3rd.
While we invite submissions that focus on any pertinent topic relevant to ancient metagenomics, this edition of SPAAM will have four sessions devoted to the following topics:
- Session 1: Ethical considerations and best practices in ancient metagenomics research
- Session 2: Current challenges and biases in ancient patho(meta)genomics
- Session 3: Urgent challenges and future steps in ancient microbiome research
- Session 4: Tool up or die - gaps and solutions in ancient metagenomics analysis
A description of the different sessions follows:
Session 1: Ethical considerations and best practices in ancient metagenomics research
Standards and expectations of responsible research are complex and culturally dependent; however, ethical aDNA research should be guided by two questions, “How does my research impact the living?” and “How does my research impact the dead?” Much of aDNA research involves the destruction of human remains from a Western scientific perspective, which is not uniformly supported across Indigenous and invested communities, specifically Indigenous groups and Tribal Nations. The degree to which invested and descendant communities can participate in the decision-making process varies widely by country, with many Indigenous groups and under-resourced communities lacking the sovereignty, scientific infrastructure, and power to control their own cultural resources. Even when researchers have support from stakeholders, it is critical to recognize that ancient remains are finite. Not only is sampling a destructive process, but also over sampling and exportation without proper documentation and repatriation plans can lead to incomplete skeletal collections, limiting future osteological, isotopic, and biomolecular research. The goals of this SPAAM session are to (1) Examine the responsibility of aDNA and laboratory researchers in addressing these concerns, and (2) Offer recommendations for responsible research participation, including best practices for ethical research design, engagement with invested groups, transparency in collaborative practices, and responsible treatment of archaeological and historical remains.
Session 2: Current challenges and biases in ancient patho(meta)genomics
Ancient remains are limited and are an invaluable part of our cultural patrimony. Thus, maximization of scientific output should be a priority in the ancient DNA (aDNA) field, given the destructive nature of the sampling procedure required by most of the studies. DNA extracted from ancient remains contains information beyond the host’s genetic component; it can also harbour pathogen DNA present at the time of the host’s death. The recovery of ancient DNA from pathogens, along with historical and archeological evidence, offers new insights into the origins, etiology, nature, and evolutionary history of ancient infectious diseases. As the number of recovered ancient pathogen genomes grows, so have concerns about the wet and dry lab strategies employed. This SPAAM session will be focused on discussing challenges and biases in the retrieval and analysis of ancient pathogen genomes. Furthermore, we would like to discuss new analytical avenues, such as epidemiological modeling or functional evolution, for ancient pathogenomics, which could provide additional valuable information for the modern microbial community. Some of the suggested topics:
- Capture experiment designs
- Authentication of ancient pathogen hits
- Variant discovery
- Phylogenetic reconstruction
- Evolutionary dating analyses
- Functional inference from mapping data
- Engagement with modern microbiologists
- Challenges in epidemiological modelling with ancient pathogenomics
Session 3: Urgent challenges and future steps in ancient microbiome research
Over the past decade, advances in DNA sequencing technologies have revolutionized the amount of data that can be recovered from archaeological materials. In particular, metagenomic techniques have been able to recover ancient microbial DNA from dental calculus, paleofeces, plants, soils, animal remains, and so forth. However, several related challenges related still remain, such as best practices for the collection of samples, decontamination, and authentication criteria. The lack of standardization makes it difficult to control for noise across datasets generated from different research groups. The goals of this SPAAM session are to (1) Address the current issues in ancient host-associated and environmental microbiome research and (2) Discuss possible ways in which the ancient metagenomic data is more reproducible and easier to compare across research teams.
Session 4: Tool up or die - gaps and solutions in ancient metagenomics analysis
Workflow and tool development in the field of ancient DNA (aDNA) has been largely focused in the study of ancient human genomics. A combination of High Throughput Sequencing and increased availability of High Performance Computing has opened the possibility of analysis of whole metagenomes. These advancements have led to broadening the types of specimens analysed, new avenues of research, and a myriad of data types being generated. Therefore only recently has ancient metagenomics come to the forefront of palaeogenomics and is primed for the development of tools to tackle the challenges of ancient metagenomics, such as, but not limited to incomplete and biased databases, validation of taxonomic profiling, authenticating datasets, contamination with modern or ancient microbes, etc. In this session we would like to (1) discuss new tools/workflows being developed for the analysis of ancient metagenomic datasets, as well as (2) review commonly used workflows and tools in the field of ancient metagenomics and how to improve them.
We aim to facilitate a collaborative process whereby early career scholars from across the globe can share and develop strategies to improve the field. We invite submissions that focus on any pertinent topic to ancient metagenomic research. These include, but are not limited to past interests:
- Best practices for authenticating ancient microbiome datasets
- Best practices for engaging with local/Indigenous communities
- Best practices for minimizing contamination and decontaminating ancient samples
- Best practices for publishing sequencing data (e.g. number of controls)
- Defining metadata standards
Submission Options
Individual splash talks
Splash talks will allow discussants to bring up ideas about a certain challenge or issue within one of the three topics. Splash talk presenters will be allotted 5 minutes to discuss their ideas. Abstracts submitted in this category may include empirical research, obstacles noted during research, theoretical development, reviews, and/or critiques. The word limit for splash talks is 250 words. Abstracts should include the following information:
- A description of the targeted audience
- Objective or problem under investigation
- Hypothesis or research goal
- Statement of why the issue is important and how can a collaborative setting can help overcome it
Collaborative Panel Presentation
Accepted proposals in this category will be organized into a panel of at least 3 related presentations. Panel presentations will be allotted 20 minutes. Abstracts submitted in this category may include empirical research, obstacles noted during research, theoretical development, reviews, and/or critiques. The word limit for panel presentations is 250 words. An abstract in this category must include:
- A description of the targeted audience
- Objective
- Problem under consideration
- Hypothesis or research goal
- The introduction of a new wet-lab or bioinformatic tool
- Statement of why the issue or tool is important and how can a collaborative setting can either improve it or benefit from it
Special Interest Groups (SIGs)
Following each panel discussion, attendants and discussants will be allowed to subdivide into SIGs, which will involve Zoom Breakout Rooms. One discussant will be assigned to a SIG while participants are allowed to join whichever one they elect to choose. Discussants will be assigned the lead role to facilitate the conversations during this period. The aim of an SIG is to foster discussions between discussants and participants to connect and collaborate in smaller settings. Following this procedure, discussants and participants will then re-convene and the discussants will be asked to summarize their outcomes with the entire group.
Accessibility
While we cannot provide financial support, we aim make the workshop as accessible as possible (babies, children, disabilities etc.). Please let us know what support you may require in the registration form under comments/suggestions.
Organising Committee
Miriam J. Bravo (LIIGH UMAN, Mexico, mbravo@liigh.unam.mx)
Aida Andrades Valtueña (MPI-EVA, Leipzig, aida_andrades@eva.mpg.de)
Elizabeth (Betsy) A. Nelson (UConn, Connecticut, enelson@uconn.edu)
Kelly Blevins (ASU, Arizona, Kelly.Blevins@asu.edu)
Sterling Wright (PennState, Pennsylvania, svw5689@psu.edu)
