SPAAM SLACK ARCHIVE
From: Oct. 2018 - Jul. 2024
Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
@Clio Der Sarkissian has joined the channel
@Antonio Fernandez-Guerra has joined the channel
@Sterling Wright has joined the channel
@channel I've just made #papers for everyone to share stuff you come across and might be useful for everyone!
If anyone is looking for a postdoc: https://evoldir.tumblr.com/post/622780427510398976/fwd-postdoc-ucopenhagenancientdna/amp?_twitterimpression=true
(ancient environmental metagenomics in Copenhagen)
Hello SPAAMers (@channel) an opportunity has fallen into my lap (/negotiated) for us to already start doing things together as a community.
My supervisor has been asked to help write a review, and a part of that she was asked to contribute a list of all published ancient microbiome samples, and I think she panicked when she realised there isn't such a list already. She then asked if I could help create this list, but I thought it was silly to make such a list just for a single (possibly paywalled) paper, when this sort of thing will evolve over time and could be useful for many different people. Furthermore, this is something having a group of people working together would make it much more efficient and last longer over time.
After a discussing with a couple of SPAAMers here in Jena, I will be making a github repository within the SPAAM organisation that will contain TSV files with these lists of samples and a small amount of metadata. This will by no means be exhaustive, but is meant to act as a reference guide for people to quickly look up where to find the sort of samples they are looking for. It will therefore be lightweight, but this comes with the benefit it will be easier to maintain, and people will therefore be more willing to add their own studies over time.
I will be setting up this repository over the weekend with some templates and start making the lists. I will also make a list of papers that people can assign themselves to get the required info from.
Finally, I will generate a Zenodo DOI for the database and we can make regular releases so each release has a stable ID that can be cited in any papers people would use the information in. I also envision that we can make a short one page 'data resource announcement' publication that can also be used for citations and CVs.
If you would be interested in helping out, please add a reaction to this message or leave a comment under the thread. Anyone who contributes (as recorded on GitHub) will be a co-author on any publication that arises.
TL;DR we will be setting up a ancient metagenome sample directory containing lists of all published ancient metagenomes. This will be citeable and updated over time. If you want to contribute (with co authorship possibilities) please let me know!
Cheers, James
*Thread Reply:* Sounds like a brilliant idea! I am not very experienced but would be very happy to help where I can 🙂
*Thread Reply:* Experienced with github you mean?
*Thread Reply:* Oh ok! I could do a tutorial Jitsi call or something.
*Thread Reply:* And then we can write documentation that 'not experienced' people can follow
*Thread Reply:* That would be great, thank you. It seems like a great chance to learn. I’m happy to help find metadata etc.
OK @channel, for those who want to get involved please see #ancientmetagenomedir for more information!
And finally, don't be put off by we are doing it on github!
If you're interested in contributing but aren't experienced with github, and want a simple how-to github tutorial - react to this message or PM me and if there is enough people I'll run a 1hr BigBlueButton Jit.si call or something to get you up to speed
@channel as a second SPAAM 'project', what would you think about having some form of bibliography of SPAAM approved publications? Assuming we agree on a set of standard data reporting, we could give a 'badge of approval' of a publication. This bibliography could then be a centralised list of where to go to look for papers where you should be able to easily find (good quality!) data from.
Do you think this would be useful? The reason I had this idea is from adding papers to AncientMetagenomeDir - some papers are really good at making the metadata data complete and accessible, but others are awful and you have to hunt through all the text and multiple SI files to get all the basic metadata. The one utility I can think of is that if this list existed, then you could quickly download with confidence a SI table from that paper without worrying about having to manually check it.
Thoughts?
I think is a good idea. I would set some type of scoring system based on a checklist that the curator can use when adding the samples to the different tables. Like does it have the minimum contextual data we require? Does it have reproducible code? Has the raw data been uploaded to one of the primary databases? Then this score can be integrated in the tables and used to generate the badge. But it should be thought/designed in a way that easily integrates in the workflow of the curator. A similar idea applied for the FAIRness of data, https://github.com/RDA-FAIR/FAIR-data-maturity-model-WG/issues/34

I like the idea too. The STROMS/STORM papers (both in the #papers channel) would be useful here to set the criteria to be in line with recommendations for modern metagenomic data
Ok, I've made a Trello-like board here: https://github.com/orgs/SPAAM-workshop/projects/1
Feel free to add/propose your own
To help us keep track of project ideas and their developments. @channel feel free to add your own proposals!
At the good suggestion from @Abby Gancz I've now made #events where people can announce events they are host/running/attending and would like others to know about!
@channel I’ve created a new #jobs channel where people can post job (PhD/postdoc/other) adverts. I just posted an advert for a postdoc opportunity in eDNA that was circulated at my institute.
Reminder, for those who will be participating in spaam2, all discussion will be happening in <#CPHECT30A|spaam2-open>
@channel Due to some slight reorganisation we have changed the github organisation to SPAAM-community. Therefore the new website will be spaam-community.github.io. The same goes for AncientMetagenomeDir.
Please be aware while we go through and update links, some functionality of the website will be a bit unstable. Please report anything that's broken. Also note those who have cloned any of the github repositories will need to update their remote URL.
Today is the day @channel for SPAAM2! Remember to please join <#CPHECT30A|spaam2-open> for the discussion channel!
could y’all please add descriptions to the channels?
*Thread Reply:* I will do a clean up after tommorrow
@channel just a reminder this is a completely open community. SPAAM =! sPAAM2, please feel free to invite anyone else you think would benefit! PIs included ;)
*Thread Reply:* Thanks James 😆
*Thread Reply:* I would like to invite someone James - I sent them this link: https://spaam-community.github.io/#/?id=standards-precautions-and-advances-in-metagenomics they said this results in Workspace not found
*Thread Reply:* Does this work: https://join.slack.com/t/spaam-community/shared_invite/zt-ei8pfw4m-XdBGTQwRaXWrEkd618YlhQ
*Thread Reply:* (it works for me?)
*Thread Reply:* I have invited them, just thought you might need to know about the dead link
*Thread Reply:* Hm that's wierd though, as that's the link on the link you sent above on the spaam website 😆
*Thread Reply:* Oh well, solved now!
*Thread Reply:* Thanks for reporting anyway 🙂
@Alex Hübner @Susanna Sabin if either of you would be interested in saving yourself time in peforming SNP filtering and table generation, please let me know and we can consider adding the steps to nf-core/eager (bit annoyed MultiVCFAnalyzer is the only option atm)
*Thread Reply:* I hope I remembered correctly who talked about that 🙏
*Thread Reply:* MUSIAL (https://github.com/Integrative-Transcriptomics/MUSIAL) works well as an alt to MultiVCFAnalyzer. It can handle all GATK3 and GATK4 VCFs, as well as those generated by freebayes. It has similar filtering functionality as MultiVCFAnalyzer and generates a SNP table. It came out of a 2019 Tübingen MSc (https://publikationen.uni-tuebingen.de/xmlui/bitstream/handle/10900/89312/Dissertation_Seitz.pdf?sequence=1&isAllowed=y). I think Alexander Herbig was even involved with dev…
I am generally interested but haven’t done it in a while. I would therefore first need some time to revise my workflow and update it. But maybe @Susanna Sabin has already something ready for you. I am very slow these days…
*Thread Reply:* zimmerframe
#events: Cycling backward: The G-BiKE workshop on ancient and historical DNA. https://sites.google.com/fmach.it/g-bike-genetics-eu/meetings-events/g-bike-adna-workshop?authuser=0 Target audience:
Overview of aDNA: general audience.
Fora: Young investigators with biological and ecological background, practitioners who want to learn more about the aDNA potentiality for conservation biology. Please, consider that you’re requested to actively participate in the consecutive sessions which are meant for promoting discussion and whose main result will be distilled into a scientific paper.
#events: Ancient Biomolecules of Plants, Animals, and Microbes (Virtual Conference) 29th- 31st March 2021 https://coursesandconferences.wellcomegenomecampus.org/our-events/ancientbiomolecules21/

*Thread Reply:* Are you sure we can trust this conference?
*Thread Reply:* Never change a running protocol
Hello everyone, I hope everyone is having a good Monday. I work with vaginal microbiota and have low coverage metagenomics sequencing data and would like to align it to human genome to see the coverage and then remove the human reads and look into remaining reads. What alignment tool or pipeline do you use for such analysis? Any suggestions would be appreciated 🙂 Thanks in advance!
*Thread Reply:* Hey Başak, are you somehow working with ancient metagenomic data? We can mostly give the best advice on ancient DNA, but can endeavour to help you regardless (even if with less expertise 😅 )
*Thread Reply:* And for the specific task that you mentioned, you could have a look at nf-core/eager, with the --hostremoval_input_fastq option https://github.com/nf-core/eager
*Thread Reply:* Was going to suggest exactly that ☝️, thanks @Maxime Borry! (although whether to run the whole pipeline or just the script itself depends if you have aDNA data)
*Thread Reply:* hi James, I'm not working with ancient DNA unfortunately. I assumed the basics should be the same 🙂
*Thread Reply:* Thanks a lot Maxime, I'm looking into it!
*Thread Reply:* For “modern” DNA metagenomics, you could have a look to https://github.com/ifremer-bioinformatics/samba or https://github.com/nf-core/mag (talking only about nf-core pipelines)
*Thread Reply:* Thanks, Deeply appreciated
*Thread Reply:* or you can try this https://github.com/SegataLab/preprocessing if you are fluent with python environment.
#events: Bioarchaeology Early Career Conference 2021 with sessions on ethical approaches to excavating human remains as well (on Day 4 Commercial and Museum). Abstract deadline 15th Jan 2021. https://becc2021.com/

@channel so I've discovered that the German Research Network (dfn.de) allows to set up mailing lists (listserv) for free for German academic institutions. The only requirement is that the 'owner' of the list must be a part of a DFN-network institution (which most members of German institutions here would be applicable). What do you think about setting up a mailing list for SPAAM, to help improve communications with non- (or less regular) slack users and for wider announcements?
Would this be something of interest, or would this be annoying? Or you dislike the idea of the mailing list having to be 'under control' of German researchers (in name only, though, note)?
Dear @channel,
As we haven’t got much feedback from you so far and are afraid that emails might have gotten stuck in SPAM folders, I will cross-post the email here again and would like to encourage you all to participate.
```Dear all,
First of all, we would like to all thank you again for your active participation in the SPAAM2 workshop in September and the vivid discussions that have since then taken place on Slack.
As it was announced in the last email, there is certainly a wide variety of directions and challenges facing our relatively young community, which can be difficult to include in one paper for a wider audience. Instead, the organisers thought that it would be more constructive to write a Q/A style FAQ opinion piece aiming to portray our field to modern microbiologists in order to reduce the chances for misconceptions and misunderstandings by colleagues who aren't working with this type of data on a regular basis.
For this, we would like to ask for your help. In order to give an accurate representation of commonly faced challenges by researchers unfamiliar with our field, we would like to ask you if you are willing to share your feedback and criticisms, both positive and negative, that you have recently encountered? We would be particularly interested in the comments by reviewers that you have received during peer review.
We are totally aware that sharing reviewers' comments is a sensitive issue. Therefore, we would like to provide multiple options how to share your reviews with different levels of confidentiality. For junior researchers such as PhD student and postdocs, please consult with your PIs if they agree on sharing the reviewers' comments prior to sending them, as this might be a delicate matter.
The first option would be to send your reviews but also general advice/feedback you regularly give as a reviewer directly via email to me. If you choose this option, it would be great if you could provided us additionally with the following information alongside your reviews:
- Would the organisers be allowed to use the reviewers' comments verbatim by copy-pasting short excerpts of about 1-2 sentences into the final manuscript or would you prefer a re-phrasing?
- With respect to data protection, could the reviews be stored on a sharing platform (Google Drive)?
In case a sharing of the reviewers' comments via email would not be possible, we would also be interested in interviewing any of you who are willing to share their experiences verbally as a second option.
Third, if you are aware of metagenomic papers with open reviews, it would be great if you could share them with us, too.
In case you are interested in contributing to this opinion piece by e.g. sharing your experiences of your review process with us, we would appreciate if you could reply to this email in the next days. We would then start to compile a list of most commonly faced challenges starting in the end of February. The deadline for sending us your feedback would be 22nd February 2021. Once we have received the responses, we will discuss how we will proceed in collaboratively writing the piece itself and how contributions will be acknowledged.
We look very much forward to your contributions.
On behalf of the organisers,```
*Thread Reply:* On it. Will get back to you in a few days.
☝️ Please also pass on to anyone else in the field who may not have come across SPAAM! Particulary senior researchers/PIs!
Hi All, I'm working with ancient, oral metagenomic communities and would like to SourceTracker analyze Metaphlan2 community profiles. Has anyone else done this? I'm having difficulty inputting the Metaphlan2 output into SourceTracker. On that note, does someone have a Metaphlan2 sources biom file? Would appreciate any help! Thanks so much!
*Thread Reply:* Hehe, ok so I'm not on childcare duty today. I just check whta I used in the past: https://biom-format.org/documentation/biom_conversion.html
However, which sourcetracker are you trying to use? The original version (not the QIIME adapted one), accepts just a TSV file: https://github.com/danknights/sourcetracker
*Thread Reply:* I have been using the QIIME adapted version since it will merge and rarefy the sources and sinks.
*Thread Reply:* Ah, then hte first link should work for you. But the R library can also rarefaction I believe (at least that's what I did, because I followed the last example on the README).
*Thread Reply:* I got MetaPhlAn2 relative abundances to work in SourceTracker and all other analyses that require count data and not frequencies by converting the relative abundances into pseudo-counts. I typically opt for 100,000 pseudo-counts to correspond to 100%, so I multiply all my relative abundances with 1,000 (relative abundance in % ** 100,000 / 100) and then work with pseudo-counts instead. No problems so far.
*Thread Reply:* which README? Would either of you mind sharing your commands? I get lost so easily 🤦♀️
*Thread Reply:* README - main page here: https://github.com/danknights/sourcetracker
*Thread Reply:* Here is the code to get the OTU table from the simple MetaPhlAn profile files.
@channel mailing list now setup! Please subscribe here (I think...)
*Thread Reply:* Seems to have worked 🙂
If a few people here in slack subscribe and this work, I'll invite the whole of <#CPHECT30A|spaam2-open> 🙂
Hi All, here’s a conference many of you may be interested in “Holistic Bioinformatic Approaches used in Microbiome Research” - it’s targeted to early career researchers (just like SPAAM2), focused on reducing the entry barrier to microbiome bioinformatics. With an emphasis on open data sharing. https://biovcnet.github.io/_pages/conference-2021/
Dear @channel, A brief reminder regarding our opinion piece update that was also sent out to our new mailing list: ```Dear all,
I would like to take this opportunity and remind you of our email sent two weeks ago asking for your help in writing a Q/A style FAQ opinion piece aiming to portray our field to modern microbiologists. In particular, we would be very interested in hearing your feedback and criticisms, both positive and negative, when faced with challenges by researchers unfamiliar with our field during peer review. Furthermore, we would like to hear what feedback/advice you regularly give as a reviewer or mentor to people less familiar with our field.
In the last two weeks, we already received some feedback but we of course would like to hear from many more of you. It was brought to our attention that the sharing of the reviewers' comments is a sensitive issue. Therefore, we would like to remind you to first talk to your PI before sharing them or rather paraphrase them for us when explaining your experience. Each experience is a valuable resource for this purpose and we would be delighted to hear your stories.
The deadline for sending us your feedback would be 22nd February 2021. So please get in touch with us if you have a moment to share your experiences with us.
On behalf of the organisers,```
Hi everyone, did anyone ever succeed in installing HOPS the way it is described here https://github.com/rhuebler/HOPS ? I spent quite a lot of time on that and can explain what exactly fails. Perhaps @Alex Hübner or @James Fellows Yates have experience running and installing HOPS?
*Thread Reply:* I would recommend installing it via conda...
*Thread Reply:* At least we use that in eager
*Thread Reply:* And it works
*Thread Reply:* Or is that how you tried to install it and it didn't work?
*Thread Reply:* I also only installed it via conda.
*Thread Reply:* Thanks guys, this is what I get when installing via conda
conda install hops -c bioconda Collecting package metadata (currentrepodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: failed with repodata from currentrepodata.json, will retry with next repodata source. Collecting package metadata (repodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: \ Found conflicts! Looking for incompatible packages. This can take several minutes. Press CTRL-C to abor| failed
*Thread Reply:* I tried it on my computer and a computer cluster, same output
*Thread Reply:* Installing it via the shell script “install.sh” can’t get the hops.jar file
*Thread Reply:* Ah shit I guess maybe Ron didn't pin some dependency...
*Thread Reply:* One sec, let me see if u can see the recipe
*Thread Reply:* I can see that the yml-file should probably be modified, for example, “getopt” should be replaced by “r-getopt” and a few other things
*Thread Reply:* Alternatively: install malt and maltextract
*Thread Reply:* And run each step manually
*Thread Reply:* (that's what we do in eager2)
*Thread Reply:* The Rscript for the PDF reports are in the repository from Felix Key
*Thread Reply:* Yes, I have malt and can install maltextract. But is this all HOPS needs? My understanding is that HOPS is a damage-tolerant version of MALT
*Thread Reply:* No HOPS is a pipeline
*Thread Reply:* Yes, a pipeline, but most of the stuff HOPS does I can probably do without HOPS. The thing I can not do is to make a damage-tolerant MALT, that is a peculiarity of HOPS as far as I can see from their publication
*Thread Reply:* MALT already accounts for damage. There is a 'cracked' version of MALT (cmalt) that has a couple of minor changes
*Thread Reply:* https://github.com/rhuebler/cMALT
*Thread Reply:* That should already have a jar file
*Thread Reply:* Post processing is here: https://github.com/keyfm/amps
*Thread Reply:* And finally: you can probably just email Ron to ask him to fix it
*Thread Reply:* huebler@shh.mpg.de
*Thread Reply:* He is still partly active
*Thread Reply:* Yes, I saw it, never tried though, but I thought HOPS should have all the cracking nicely packed together, did not expect it to be a headache to install HOPS
*Thread Reply:* Thank you, I opened an issue on the github, but will also write to Ron
*Thread Reply:* Yeah so hops itself was originally built with slurm integration but the reviewers of the paper (I believe) requested something a bit easier. So maybe conda recipe was done a bit 'fast'
*Thread Reply:* James, does eager2 install HOPS? Or cMALT? If HOPS, how does it do it if the conda installation is broken?
*Thread Reply:* https://www.github.com/nf-core/eager/tree/master/environment.yml
*Thread Reply:* Huh ok, apparently hops!
*Thread Reply:* I suspect it's because we pin the Java version
*Thread Reply:* (the source of most problems...) So if you specify you want to specifically install openjdk 8 alongside the hops package with conda,I guess it will work
*Thread Reply:* Thanks, I will try it!
*Thread Reply:* I had all kinds of trouble installing HOPS earlier in the pandemic-- I wrote to Ron and he sent me a link that was as of March 2020 unreviewed, but worked:
conda install -c <https://99494-42372094-gh.circle-artifacts.com/0/tmp/artifacts/packages> -hops
*Thread Reply:* This worked once I created a new environment for HOPS and then used the above install command:
conda create -n hops_test
conda activate hops_test
conda install -c <https://99494-42372094-gh.circle-artifacts.com/0/tmp/artifacts/packages> -hops
He said review times were of course running slow due to the pandemic, hence passing me this version that “passed all tests” but had not yet been reviewed and therefore was not yet available at the download link. However I imagine that has since changed
*Thread Reply:* I’ve never had luck running the entire MALT-HOPS pipeline in one go though--I have to run MALT and then run HOPS on the output in mode me_po
*Thread Reply:* Oh that's horrible 😱
*Thread Reply:* @Nikolay Oskolkov please let me know Ron said it's still under review. I know a few people on bioconda team
*Thread Reply:* So can expedite the process
*Thread Reply:* I guess if my link works and the download still doesn’t, then they could probably use some expediting… but this was almost a year ago!
*Thread Reply:* Thanks Shreya, are you sure there is “-hops” at the end of “conda install ...” line?
*Thread Reply:* I mean, it should probably be just “hops”, but anyways the command line you posted gives me “UnavailableInvalidChannel”
*Thread Reply:* It should just be “hops”, but I think if the link doesn’t work then it’s probably out of date 😕
*Thread Reply:* I’d definitely write to Ron in that case! He was very helpful to me
*Thread Reply:* James, specifying openjdk=8 in the yaml file did not help either, hopefully Ron can fix this :(
*Thread Reply:* Does it report the exact conflict?
*Thread Reply:* Can you get a verbose output
*Thread Reply:* After some tweaking, the following yml-file worked for me:
Create environment:
conda env create -f environment.yml
name: HOPS channels:
- conda-forge
- bioconda
- defaults dependencies:
- conda-forge::python=3.7.3
- conda-forge::openjdk=8.0.144 # Don't upgrade - required for GATK
- bioconda::hops=0.34
Then I run: conda activate HOPS
*Thread Reply:* and it seems to be working since "hops -h" prints:
hops -h usage: HOPS [-c <String>] [-h] [-i <String>] [-m <String>] [-o <String>] [-v] HOPS version0.33 -c,--configFile <String> Path to Config File -h,--help Print Help -i,--input <String> Specify input directory or files valid option depend on mode -m,--mode <String> HOPS Mode to run accpeted full, malt, maltex, post -o,--output <String> Specify out directory -v,--version Print Version In case you encounter an error drop an email with an useful description to huebler@shh.mpg.de
*Thread Reply:* Thanks a lot @James Fellows Yates and @Shreya for your help!
*Thread Reply:* Do you have an idea what fixed it?
*Thread Reply:* It might be worth checking the hops recipe to see if you can fix it
*Thread Reply:* Previously, I cloned the hops repository and used the yml-file that was in the "Installation" folder. The yml-file looks like this:
name: hops channels:
- bioconda
- anaconda
- conda-forge
- defaults
- R
dependencies:
- openjdk=8
- R
R
- parallel
- r-getopt
- r-gridbase
- r-gridextra
So I created environment "hops" with this yml-file, activated it and ran "conda install hops -c bioconda" within the environment. The idea was that openjdk=8 was pinned in the environment. This way did not work. Then I simply took the yml-file from eager that James posted and apparently this line "- bioconda::hops=0.34" was what made it work. Not sure whether it is the version 0.34 that should have been specified or just it is better to install hops together with creation of the environment
*Thread Reply:* So, in summary:
conda install hops -c biocondaas it is suggested here https://github.com/rhuebler/HOPS, does not work- conda env create -f environment.yml with the following yml-file:
Create environment:
conda env create -f environment.yml
name: HOPS channels: - conda-forge - bioconda - defaults dependencies: - conda-forge::python=3.7.3 - conda-forge::openjdk=8.0.144 # Don't upgrade - required for GATK - bioconda::hops=0.34
Worked for me
*Thread Reply:* I wonder if need to be more precise with the haha version in the hops recipe...
*Thread Reply:* Ok, running "conda install hops=0.34 -c bioconda" in the base environment or in an empty environment created with "conda env create -n myenv" gives me: Collecting package metadata (currentrepodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: failed with repodata from current_repodata.json, will retry with next repodata source. Collecting package metadata (repodata.json): done Solving environment: failed with initial frozen solve. Retrying with flexible solve. Solving environment: | Found conflicts! Looking for incompatible packages. This can take several minutes. Press CTRL-C to abort. failed
UnsatisfiableError: The following specifications were found to be incompatible with each other:
Output in format: Requested package -> Available versions
But when I create a test environment with the following yml-file:
Create environment:
conda env create -f test_env.yml
name: test_hops channels:
- conda-forge
- bioconda
- defaults dependencies:
- conda-forge::openjdk=8.0.144 # Don't upgrade - required for GATK
Then running "conda install hops -c bioconda" works!
*Thread Reply:* So @James Fellows Yates I think you were right that it is important to create an environment with "openjdk=8.0.144" fixed (even though I actually did "openjdk=8" when using the yml-file from the cloned hops repository)
*Thread Reply:* I think it is not the version 0.34 of hops but "openjdk=8.0.144" that did the job. Mysterious!
*Thread Reply:* Ok, maybe you can can tell Ron and he can update the recipe
*Thread Reply:* Should be simple for him
*Thread Reply:* Yes, will send this to him!
Related to the thread above 👆 Qu.: Has anyone managed to get the HOPS pipeline to work with a PBS queuing system? It’s adapted to slurm, so I’m having to execute the commands separately within the conda environment, which works fine, but would be nice to have the pipeline function. I will ask Ron as well, but just wondering if anyone already has it working with pbs by chance 🙂
*Thread Reply:* Sorry @Åshild (Ash), we have slurm on our cluster, so I can't say about pbs, but I have not even tested the slurm function of HOPS yet. So far I am using HOPS in a conda environment as you do. Btw, may I ask if you managed to produce sam-alignments with HOPS, i.e. not only the rma6-file?
*Thread Reply:* You could have a try at running it through eager
*Thread Reply:* Nextflow natively supports PBS Torque https://www.nextflow.io/docs/latest/executor.html#:~:text=PBS%2FTorque,The%20PBS%20executor&text=of%20batch%20schedulers.,Nextflow%20manages%20each%20process%20as%20a%20separate%20job%20that%20is,scenario%2C%20the%20cluster%20login%20node|https://www.nextflow.io/docs/latest/executor.html#:~:text=PBS%2FTorque,-The%20PBS%20executor&text=of%20batch%20schedulers.-,Nextflow%20manages%20each%20process%20as%20a%20separate%20job%20that%20is,scenario%2C%20the%20cluster%20login%20node.
@channel There is now a draft programme for the Wellcome trust conference "Ancient Biomolecules of Plants, Animals and Microbes", I'm posting on #events the sessions that might be interesting to SPAAMers
Apologies for posting here instead of in <#C01B7CRJK7U|seda-dna>, but there aren't many people in <#C01B7CRJK7U|seda-dna> yet 😄
Does anybody know someone with experience of (preferably) shotgun (definitely) sedimentary aDNA that would like to try being an external examiner? The sedaDNA community is rather small at the moment...
*Thread Reply:* Hey Becky, have you thought of Laura Parducci? Her current PhD student Kevin Nota is doing amazing work on plant material from lake sediments.
*Thread Reply:* Thanks Katerina, but we've already had to rule out Laura. Maybe Kevin would be up for it in a few years though!
*Thread Reply:* If you've not already, although it might take a bit of time you could go through the SPAAM attendee list and Google each person to see what they work on and who their supervisor is?
*Thread Reply:* Although I'm assuming you know about this already, there is this you could look through: https://ercapo.wixsite.com/sedadna-society
*Thread Reply:* Already checked SPAAM, but I was woefully ignorant of the society. Thank you!
*Thread Reply:* Thanks everyone - someone we'd assumed didn't want to got back to us today saying actually they would, so we've found an examiner! But I have passed these lists on so hopefully we won't be so desperate again :)
Congrats to @Sterling Wright for his newly awarded grant!
https://twitter.com/Sterling1Wright/status/1365749612494077953?s=19

Hi, @channel! Check out SPAAM’s fresh new twitter account! While you’re there, fill out this poll to indicate when you would like SPAAM3 to be held.
Which week would work best for you to attend three consecutive days of 4-6 hour sessions of ancient metagenomic goodness?
@channel please also let us know in #papers if you have a paper published, then we can also tweet it on the Twitter account!!
Does anyone know if anyone has ever properly done a 'benchmarking' study, to show how short reads can really go with k-mers and it still sort of works?
@Maxime Borry @Nikolay Oskolkov might know of anything?
*Thread Reply:* James, as far as i remember in the very first Kraken paper they justified their choice of default kmer equal to 31 as most of the organisms can be reliably distinguished with 30nt kmers
*Thread Reply:* I think there are studies (sorry don't have refs so far) where people trained Naive Bayes classifiers to discriminate between different organisms and concluded that 30nt is a realiable enough sequence length
*Thread Reply:* Ok this is interesting because maybe they do work for aDNA
*Thread Reply:* Despite people saying they don't
*Thread Reply:* yeah, good question about aDNA. In evolutionary science as far as I know, we can use 30nt sequence to quite reliably assign it to an organism (providing that the complexity of the sequence is satisfactory, i.e. it is not a repeat), but whether 30nt is good enough for a damaged sequence, I do not know
*Thread Reply:* Yeah that's my remaining question, I'm not sure how classifiers are with that (maybe you could modify the seed sequence which some use). On the otherhand, damage is only at a frequency e..g. 30% so maybe it's still OK?
*Thread Reply:* damage is not so important as the length of the sequence
*Thread Reply:* My understanding is that a damaged 30nt long sequence will not be classified at all. However, reads longer than 30nt may be damaged and they can still be classified. This is because a read is split into multiple 30mers and each 30mer "votes" for an organism from a database. This means, the 30mers containing damaged nucleotides will not "vote" for any organism (or "vote" for a "wrong" organism) but the majority of 30mers coming from non-damaged parts of the sequence will still vote for a "correct" organism, and the majority vote will still assign a read to a "correct organism"
*Thread Reply:* we have been testing this, for example if you use kraken, you need to tune the DB building
*Thread Reply:* we model first the sequences we want to classify, and then tune the DB building based on this
*Thread Reply:* For example, I know for sure (I tested) that reads with adapters present are still ok to feed to Kraken, they become correctly classified. This means that some "noisy" nucleotides do not influence classification that much
*Thread Reply:* we are going to be releasing soon a full workflow of db building and benchmarking for modern and ancient DNA
*Thread Reply:* using default DBs for short reads like aDNA reads is not a good idea
*Thread Reply:* This is satisfying to know @Nikolay Oskolkov! Because then my understanding of the system is correct 😆
*Thread Reply:* @Antonio Fernandez-Guerra by modelling you mean te k-mer distibution?
*Thread Reply:* different parameters
*Thread Reply:* k-mers, read lengths, damage
*Thread Reply:* then we build synthetic data sets based on this to tune the DB buikding process
*Thread Reply:* > using default DBs for short reads like aDNA reads is not a good idea In what way? The k-mer distributsions (or hash-maps, or w/e) are 'polluted' with stuff that could result in false positives or something?
*Thread Reply:* similar happens with sourcetracker, we are also developing a new version of meta-sourcetracker that takes in account all the aDNA singularities to create proper sources
*Thread Reply:* @Antonio Fernandez-Guerra so you are building a "project-specific" database that matches your sequencing data?
*Thread Reply:* But that requires pre-knowledge what you want then, right?
*Thread Reply:* Sounds cool though 😄
*Thread Reply:* @James Fellows Yates this is how we design e.g. a MALT database, we build it in a project-specific way
*Thread Reply:* So you say you are specifically looking for species: X, Y, Z?
*Thread Reply:* in our case we model the characteristics of the data
*Thread Reply:* so we are more in the way of what are the best parameters to classify sequences like this
*Thread Reply:* we don’t care who is in the pot so much as we do full ecosystem reconstructions
*Thread Reply:* What we do, we screen our samples (thousands) with KrakenUniq and select most reliable (using breadth of coverage as a criterion) organisms present in at least one sample, and use those organisms to build a MALT database for further aligning the data with MALT
*Thread Reply:* Ok cool,that's what I'm also intersted in @Antonio Fernandez-Guerra.
*Thread Reply:* Ah ok @Nikolay Oskolkov so you do generic pre-screening then validate with MALT after. Understood 👍
*Thread Reply:* we use many similar concepts that the ones described here https://instrain.readthedocs.io/en/latest/important_concepts.html
*Thread Reply:* @James Fellows Yates Exactly! We do not use MALT for screening because of many reasons (it is too slow, has a limited size database, needs lots of RAM), so we screen with KrakenUniq against the full NT and use MALT for validation
*Thread Reply:* especially for the DB construction
*Thread Reply:* OK good to know I'm not alone in going in this direction then.
And looking forward to seeing the outcome of that project @Antonio Fernandez-Guerra , sounds exciting!
*Thread Reply:* we are trying to improve what you can get from mt-sourcetracker and it also works nicely to get better taxonomic annotations
what does can really go with k-mers mean?
Like how short a k-mer can go before it becomes mostly 'uninformative'
(I know it's a gradient, but if there is a rough feeling)
you might want to read this https://www.nature.com/articles/srep28840
*Thread Reply:* it has refs that can be useful like https://link.springer.com/article/10.1186/gb-2009-10-10-r108

*Thread Reply:* these ones also might be of interest as it relates k-mers with species boundaries https://academic.oup.com/mbe/article/34/10/2716/3976054 https://www.biorxiv.org/content/10.1101/569640v5
*Thread Reply:* I'm actually interested in non-prokaytotes but thanks!
*Thread Reply:* i am quite biased on this, but the basics should be the same
@James Fellows Yates I was hearing to the talk at the bottom of this message and there they sort of tested it for the 1My mammoth at their conclusion was that below 35-30 it does not really work. I remember that Simon Rasmussen also tested that computationally and as a conclusion of his tests he was also using a min length of 30 bp. https://www.youtube.com/watch?v=LOVdbayRBns
So, if in the most challenging case so far, where every molecule would have helped, they still decided to stay above 30bp is probably because it really gets noisy below that threshold
but this is read length not k-mer length
I thought James question was for both read length and k-mer size
I didn’t understand his question when he did it so most probably i am wrong… 😅
@James Fellows Yates k-mers… you should check the strobemers https://www.biorxiv.org/content/10.1101/2021.01.28.428549v2
Sorry basically both - at what read length does your k-mers become uninformative. Everyone touched on the stuff I wanted to know, don't worry!
@Nico Rascovan you are right, in the supplementary information for the paper on the 1.6 mln years old Krestovka mammoth, Love Dalen and the guys say they used some iterative approach to determine a minimum read length that they could trust, i.e. below this read length they observed spurious mapping of bacterial reads to the elephant genome. This minimum read length was sample specific but was typically around ~35bp
*Thread Reply:* The leipzig group at EVA also showed this. Going lower (e.g. 25bp) only works when you have exact SNPs they were interested (in well studied neanderthals, so doesn't really help here)
you can use this to test this: https://www.biorxiv.org/content/10.1101/611160v1.full
you can look at genome mappability
we use this to break similar genomes in discriminative regions, but you can use it for example to test where a certain k-mer will map in a collection of sequences
One former staff scientist at the MPIEVA once more or less showed with a back of the envelope calculation that it doesn’t make sense to map reads shorter than 24 bp because the Burrows-Wheeler algorithm would be inconclusive. So most studies stopped at either 30 or 35 bp. For k-mers, I think it depends on your question of research and is highly dependent on genome mappability as pointed out by @Antonio Fernandez-Guerra above. The low hanging fruit answer is: just simulate it and see! 😜
The problem, in part, is the endogenous DNA content. Below ~35 bp you always get a random-mapping component and this is exacerbated with poorer endogenous DNA content. Krestovka had the lowest endo. DNA content of the three old mammoths. This effect can be clearly seen in the fragment length distributions in Extended Data Figure 3. We went with a global 35 bp minimum across all specimens for analytical consistency. During data exploration, we also got some weird d-stats results when including the 30-35 bp reads (again probably from spurious mapping artifacts).
We just heard a similar story from our collaborators working with fecal samples: When studying the host genome (endogenous content <1%), it was the mapped read length that was most important. The fragments were long, but everything below the mapped read length of 35 bp was unreliable and most of it resulted from cross-mapping of bacterial reads to the host genome
Does anyone know of published data with internal barcodes (or have some unpublished they don't mind us using/subsetting+anonymising for testing of nf-core/eager)? We need as much 'variety' of library construction strategies as possible to make sure our implementation works
*Thread Reply:* What is the state of the uploaded data? DOes it have index+adapter+barcode? Or just barcode?
And do you have the barcode list in the paper?
*Thread Reply:* demultiplexed+adapter removed&quality-trimmed. Internal index (barcode) still there
*Thread Reply:* I’d have to find the list, but I won’t get to it today, I can do it tomorrow
*Thread Reply:* They’re 6bp and you can see them in the FastQC report
*Thread Reply:* Wonderful thank you
*Thread Reply:* Almost all of our data is with internal barcodes, some published, some not yet. Happy to share
*Thread Reply:* Does this have what you need? I can’t find the index sequence for CS05 (there was a lot of confusion about the correct internal index list at the time)
*Thread Reply:* That's perfect @ivelsko!
@Katerina Guschanski hehe I was hoping you would say that ;). If you could somehow supply e.g. 3 unmerged/untrimmed libraries with adapters and internal barcodes with the barcode list, subset to just the first 10,000 reads that would be amazing. I can also do the subsetting if you need.
*Thread Reply:* Oh and also the ERR code for each library so I can cite in the test data documentation
*Thread Reply:* Do you dual barcode? Or just single?
*Thread Reply:* Gonna ping @Jaelle Brealey here in the hope that she'll be able to supply what you need. Wonder if the published data has what you need. If not, Jaelle, I'm happy with sharing the unpublished sequences for the AMR study
*Thread Reply:* Dual barcode, dual indexing
*Thread Reply:* Meaning 2 internal barcodes and 2 indices in the Illumina adapters
*Thread Reply:* That is perfect!!!
*Thread Reply:* I think ENA requested that internal barcodes were hard-clipped from the 5' end of the reads before uploading, so the published data won't be appropriate. I'll check the data from the unpublished study and get back to you in the next few days 🙂
*Thread Reply:* Yeah ok, that's what I expected as they don't want technical artefacts.
Thank you very much!
*Thread Reply:* @James Fellows Yates I can send along the raw fastq's for a few of our samples today -- ones with dual indices and internal barcodes. Also have the barcode and index information in a spreadsheet. How do you usually set up transfers? Do you want to DM me the details :)
*Thread Reply:* Hi Adrian! That would be great!
That all sounds great! DM is fine. And I don't mind how you ship it - I'm flexible , whatever is easier for you!
For our members who aren't on twitter: You can never have enough tools in your toolkit! We are considering adding a session to SPAAM3 that will focus solely on new and emerging tools and how to use them. Is this a session you want to see as part of SPAAM3?
Congratulations to @Anneke ter Schure!
https://twitter.com/Anneke_tSchure/status/1376195685272408070?s=19
@channel Hey, party people! In case you missed the email and tweet, #SPAAM3 is coming to a screen near you on September 1-3! Submit your abstracts and register here by May 21st: https://tinyurl.com/4a9s7jr4 We’ll palaver about pathogens! We’ll gush about microbiomes! We’ll get hyped about up and coming tools! And, importantly, we’ll discuss how to design and practice ethical research! Check out the SPAAM website for all of the juicy details.
*Thread Reply:* @Kelly Blevins I just looked at the website and it has September 2020.
*Thread Reply:* I fixed it, but it hasn’t updated yet
Hello @everyone!
I wanted to ask your opinion: if theoretically a summer school in ancient metagenomics were to be offered, what would be the most pressing thing you would want to get out of it/learn?
This can be big or small things, and generic or specific (i.e. how to use GitHub, to I want to understand why XYZ happens because of damage etc.).
You can either reply here/PM me privately, or use this anonymous Google Form (https://docs.google.com/forms/d/e/1FAIpQLSfDxuuubEzkbUcAJsU-J4sXGVvuxQOwOpX5bvC5X78nw1cjFA/viewform?usp=sf_link)
Cheers,
(also it can be for all aspects: microbiome, pathogens, env. DNA etc. etc.)
I've just made a channel called #sampling If you want to ask for help, advice, or generally discuss on how to sample things (teeth, bones, sediment w/e) for ancient metagenomics!
(remember to click on the channel name above to join, otherwise you won't see notifications)
Hi all, there is a talk happening today (may have already happened actually..I'm in Canada) by Maria Spyrou on the Black Death that I was hoping to watch, but I forgot to register for the conference. If anyone is attending and could give me the "Coles Notes" I would really appreciate it 🙂. You can PM me on here. Thanks all!!
*Thread Reply:* Maybe @Maria Spyrou can help!
*Thread Reply:* Hey Jessica, sorry for replying so late! Happy to discuss more via email 🙂
Hi! Quick question about SPAAM3 because I got confused while going through registration: is it still possible to submit abstracts and/or are more wanted? The website said the deadline was on the 21st of May but the despite clicking no abstract the rest of the registration process is pretty speaker-focused. Is that because it's still open? I'm not a confident metagenomics practitioner, but if there's a space in the conference where as beginners we can briefly talk about our current research and maybe get input from others on good guidelines/analysis I could get my courage up to present my sloth coprolite work. However, I also feel that the slack channels may be a good venue for that, rather than the conference itself? Also, let me just say I really enjoyed SPAAM2 and am really looking forward to attending SPAAM3!
*Thread Reply:* Hi!! The submission for abstracts is still open and we are happy for more submission! SPAAM3 is thought as more of a discussion conference if that makes sense, so beginners are more than welcome to present their work and the challenges they are facing. We are Looking forward to your submission :) and if you have any other questions let me know!
So to clarify: deadline in application form is WRONG at the moment. See emails/tweets.
And overall most spaam s are designed to be discussion workshops, so if you want to get feedback (not just present results of finished publications, although this is also ok) you should also submit!
If anyone is regularly using MALT (or MEGAN), I've made a bash wrapper script for a command-line way of going from rma6 ➡️ OTU table without having to open MEGAN (I got fed up having to download all my RMA6 files to my laptop or set up megan-server each time...):
Feedback gratefully received
https://github.com/jfy133/rma-tabuliser
*Thread Reply:* Thanks a lot James! Wouldn’t MaltExtract run on the RMA6 files produce a similar count table of microbial abundances?
*Thread Reply:* No, because you have to specify a taxon list to MaltExtract, and then you only get results for those taxa
*Thread Reply:* The new script allows you to do it without prior knowledge what is in your samples
*Thread Reply:* Ah, I see, interesting! I would guess you might get too many microbes then, and majority will probably be false-positives, because I guess you can't really check the breadth of coverage on the fly when generating the count table?
*Thread Reply:* I indeed quantify microbial abundances (with MaltExtract) for a specified list of taxa that previously passed breadth and depth of coverage filters
*Thread Reply:* Oh, I just thought, you actually know what taxa were included into the MALT database, so you can provide this full list list to MaltExtract, and get a count table that should be equivalent to the one obtained by rma-tabuliser, is my understanding correct?
*Thread Reply:* 1) yes correct, but depends on your purpose ;) 3) yes at a fundamental level you could, exactly. However rma-tabuliser has a few extra functionalities derived from Megan e.g. export taxids rather than names, export all assignments at higher taxonomic levels (so you don't have to manually make higher level name lists), and you can use it for functional teams tables as well (kegg, seed etc) that you can also get from rma6 files
*Thread Reply:* In other words is more lightweight and focuses purely on whereas maltExtract has all the extra authentication stuff
*Thread Reply:* Purely on ount tables
*Thread Reply:* Ach fuck slack
*Thread Reply:* You get the point
*Thread Reply:* Right, I think I got it! Thanks a lot for sharing James and I will try to test it. Currently, I am quantifying microbial abundances from RMA6 files with MaltExtract but would be curious to compare this way with rma-tabuliser 🙂
*Thread Reply:* Yeah might make more sense in this case, particularly if you want to compare e.g. genus abundances rather than species, for example
*Thread Reply:* You should get approximately the same output, but I expect rma-tabuliser to be faster (not confirmed though)
I also step on that pain when using megan before
Does anyone know of any paper which had problems or difficult to analyse because the sample was excavated e.g. in the C18th or had incomplete or sparse collection information?
Did anyone else get an email e.g.:
If so PLEASE IGNORE. I don't know what this is
Apparently someone from Universidad La Laguna? Maybe someone had struggling subsrcribing? (@Javier González Serrano would you happen to know?)
*Thread Reply:* I don't know what that is either! I am properly subscribed and did nothing to promote it (at least intentionally)
Please give your opinions!
https://twitter.com/jfy133/status/1423564438859288579
@channel the SPAAM3 (<#C025AP39256|spaam3-open>) commitee made the point the code of conduct should also apply outside of events, and therefore we will be making a SPAAM wide one.
We would need a couple of volunteers to act as contact points for anyone who needs to report . Ideally this would be with gender, instiute, and career level parity. Please PM if you would be willing to do so (it would just require having your name and email listed at teh bottom of the CoC)
Hey, @channel! SPAAM3 is just around the corner on September 1-3! Here is the complete program, one in PT and one in CEST. You can also find the program on the SPAAM-community website. Love what you see but didn’t register? We still have a few slots left. Contact one of the SPAAM3 organizers to register and attend!
There has been a proposal to set up a SPAAM blog, for short-form pieces that aren't big enough for full on papers but still useful information to learn about (e.g., what exactly is the 'theta' parameter in aDNA damage estimates, or 'should I and how can I upload my blanks to the ENA'). This would be open to any SPAAM member to post there.
I would suggest hosting it on the SPAAM website (e.g. spaam-community.github.io/blog), and it would be written in Markdown (I would write some brief instructions on how to do this, but it's quite straightforward)
My questions to the community @channel:
1) Who would be interested in reading, or even contributing to such a blog? 2) Any suggestions of what you would like to read on such a blog? 3) Are you happy that it would be hosted on our website, or would you suggest somewhere else? 4) Any name suggestions for such a blog (otherwise I'll just call it SPAAM blog)?
*Thread Reply:* Looking at you @Anna F. for name suggestions 😉
*Thread Reply:* Just for clarification: “our” website is spaam-community.github or mpi-eva?
*Thread Reply:* spaam-community.github of course!
*Thread Reply:* Would be super interested in reading it! Think this is a fantastic initiative!
*Thread Reply:* As a novice on the topic, I would love to read more accessible pieces on terminology, methods, etc, something that would make it easier to read full papers and understand conference presentations!
*Thread Reply:* I also like the idea and would read it
*Thread Reply:* I would also read it! Might even be easily convinced to write something for it.
In addition @Marcel Keller suggested having another page along the lines of 'announcments' - such as for publications by SPAAM members, and/or upcoming relevant conferences. Would people be interested in this too?
*Thread Reply:* This is very nice too. I don't think it will necessarily need someone in charge of this. It could be just done by the community itself: each time someone wants to announce something, it can just do it by following a set of instructions indicated in the blog itself
*Thread Reply:* Wonder, do we need that on top of the Slack channels? Is it primarily targeted to people not on Slack?
*Thread Reply:* the blog would reach a broader audience, perhaps not everyone wants to join the slack channel especially if only interested in some aspects
Note these would require volunteers to help monitor and maintain! So if everyone just says 'I would read it' would be insufficient. Such maintainer roles normally only takes up a couple of hours a month.
*Thread Reply:* I am happy to do some monitoring and maintenance
*Thread Reply:* Ok, maybe we can find other volunteers as well in SPAAM3
*Thread Reply:* Love the blog idea and tentatively down to help maintain, if I could learn more about it!
*Thread Reply:* Would be relatively straight forward! Just checking submissions 🙂
*Thread Reply:* I'm also happy to help monitor and maintain!
I think it is a great idea. It will be very useful for many! 1) I am sure people in my team will use it (including me), 2) I think it could be divided in sections, similarly to how the slack channel is done, to keep it organized, if there's time in the SPAM meeting this week, the structure and sections could be discussed live 3) where is hosted is not an issue I think. The biggest need would be to be able to assure continuity and enough storage, 4) If there is an interest in giving a wider recognition to SPAM as a community and entity, I think "The SPAM Blog" sounds great.
*Thread Reply:* Good idea
@aidanva @Miriam Bravo @Kelly Blevins @Betsy Nelson do you think there would be time/scope to talk about this during SPAAM3?
*Thread Reply:* We could discuss it on the last day, after the tools session
Hi @channel!! SPAAM3 is starting today!! If you are taking part and you are still not in the <#C025AP39256|spaam3-open>, join!!!
We are looking for volunteers to participate in a one day 'metadata-cleaning-a-thon' for #ancientmetagenomedir .
This would involve reading various ancient metagenomics papers, cleaning up some pre-prepared data, and filling in some gaps of *library-level* metadata referenced in the the publications (i.e. for FASTQ files). This will allow tools to be developed to download and valiate data for you (e.g. imagine one command to download all ancient Y. pestis data, or all ancient calculus).
If you're interested in helping out, please indicate your availability in October or November with the following doodle (please use full names, so I can follow up).
https://doodle.com/poll/cye42azbv77xxbeb?utm_source=poll&utm_medium=link
*Note all times are in CE(S)T, but we would start earlier/finisher later for other time zones as necessary*
Please contact me if you would like to more information (or help convincing supervisors etc ).
*Thread Reply:* @Betsy Nelson @Miriam Bravo @aidanva @Abby Gancz @Shreya @irinavelsko @Alex Hübner @Lucy van Dorp @Marcel Keller
Ping you guys as you've shown some interest. I decided after talking to a couple of people having such a day would be the most efficient to get it 'done'
*Thread Reply:* @Maxime Borry
@channel (sorry for the pinging spam) as incentive , @Maxime Borry and I have come up with a plan on how we could have a publication out of it.
During the one-day metadata-a-thon, while we compile the metadata, Maxime is going to develop a tool to do the downloading (and downloading QC) for you.
e.g. imagine the following: ancientmetagenomedirtools download --table singlegenome --species yersinia_pestis --country germany
Anyone who contributes to compiling the metadata would be included in the publication of the tool. So please sign up on the doodle to indicate availabliliy!
note you do not have to have contributed to original 'Dir publication! Newcomers are welcome!
*Thread Reply:* Everyone is also very welcome to contribute to developing the tool as well 🙂
*Thread Reply:* @Katherine Eaton if you're still working in the field, maybe something you'd be interested in ? 😉
*Thread Reply:* How much genetics knowledge is required for this? If it's relatively simple data-entry, I can help if needed.
*Thread Reply:* If you can read a paper and find keywords it should be relatively straight forward!
*Thread Reply:* We can also pair people up in mentor-mentee systems
*Thread Reply:* The more the merrier!
*Thread Reply:* I’d really like to help! But I’m on NYC time so I’m wondering if there’s any way I can do a second ‘shift’ sort of thing (like starting at 7am my time instead of 3am). Since I’m new to it all it I understand if that doesn’t work for getting up to speed, but thought I’d ask!
*Thread Reply:* Hi @Olivia! Please still sign up, it would be great to have you!
There are already a few others also in N. American who have signed up, including some AncientMetagenomeDir experts.
I will also plan to continue for a bit in the evening my time as well so can help initiate the second shift and get everyone up to speed :)
*Thread Reply:* Would love to help if I can, I am in Brisbane Australia time zone
*Thread Reply:* @s.wasef please still sign up, depending on how many people from each time zone sign up we can adjust accordingly 🙂
*Thread Reply:* Intrigued by helping @Maxime Borry with the tool!
*Thread Reply:* @James Fellows Yates, I'd be keen to be involved as well if it's not too late! Sorry, only just joined the slack today. Let me know if you can still use someone. I'm based in New Zealand.
*Thread Reply:* Oops, sorry missed this thread sorry @Meriam van Os! Will send you the detials. Do you have a google account? I can then send the google calendar invite. I also saw you sent your github account, will add you to the repo now
@channel last call for the doodle poll ☝️ (I will shut it tomorrow) If you want to help with the next stage of ancientMetagenomeDir please sign up!
For each time zone I will do introductions to the whole thing, so don't worry if you've not got involved before!
I've just made a new release of my R package for visualising the endogenous content of ancient microbiome samples (used to help you filter for preservation): https://twitter.com/jfy133/status/1437742951237627904
Very reassuring when even the authors make typos in their own packages 🙂
*Thread Reply:* Just makes you a bit more human 😂
*Thread Reply:* At least the command is right!
*Thread Reply:* > Just makes you a bit more human Shh don't tell anyone 😉
*Thread Reply:* 😶
*Thread Reply:* He does it on purpose so we THINK he's actually human 😆
As we've had a quite a few new people joining in the last week or so, here is a reminder of some channels/projects we have going on:
#ancientmetagenomedir - the first SPAAM community-wide project to aggregate and standardise ancient metagenomic sample metadata (and soon to be libraries!) <#C02DCKJ54JX|no-stupid-questions> - judgement free place to get advice and help on most basic questions when it comes to ancient metagenomics (useful for new people!) #papers - sharing of relevant paper to the field #jobs - channel for sharing relevant job postings (PhD, postdoc, group leader positions etc.) #sampling - advice on sampling techniques <#C01ASL44AMV|it-crowd> - channel for discussing the more nitty gritty in the bioinformatics when it comes to ancient metagenomics <#C02D3DJP3MY|spaam-blog> - a new upcoming project to have a blog that provides short, accessible but useful information, tips and tricks on things you vaguely know but want to know about the details on (and more!)
And finally the various SPAAM workshop channels
Hello, has anyone ever tried to directly compare the same set of ancient dental calculus samples using 16S amplicon sequencing and shotgun sequencing?
*Thread Reply:* Ziesemer, K. A., Mann, A. E., Sankaranarayanan, K., Schroeder, H., Ozga, A. T., Brandt, B. W., Zaura, E., Waters-Rist, A., Hoogland, M., Salazar-García, D. C., Aldenderfer, M., Speller, C., Hendy, J., Weston, D. A., MacDonald, S. J., Thomas, G. H., Collins, M. J., Lewis, C. M., Hofman, C., & Warinner, C. (2015). Intrinsic challenges in ancient microbiome reconstruction using 16S rRNA gene amplification. Scientific Reports, 5, 16498. https://doi.org/10.1038/srep16498
?
*Thread Reply:* Super Jamie at rescue, once again!
*Thread Reply:* If the world was full of James it would certainly be a better place
*Thread Reply:* Although the article mostly answers to my question, I still wonder if differences in beta-diversity analyses would follow a similar trend between 16S and shotgun, despite the big differences in the taxonomic profiles of both approaches
*Thread Reply:* But the point is you can't get a good estimate of (ancient) diversity though, because of the skew?
*Thread Reply:* I see what you mean. But imagine there is a group of samples that is more similar to each other than to a second group of samples. Would we see this clustering anyway, even having the huge 16S bias?
*Thread Reply:* We did that here too
But I’m not sure if it answers your question as each group was treated differently.
Also we didn’t compare them to other groups and so I can’t answer your question about beta diversity
*Thread Reply:* Thanks Sterling! Yes, I had this paper in mind. I will re-look at it with my question in mind
*Thread Reply:* Sounds good if you need any help.
Hey all, my name is Merlin, I was invited by Maxime to the Slack 🙂 To introduce myself: I life in Leipzig and I'll start my PhD at the MPI EVA in october. I am currently working on a nextflow pipeline for the analysis of captured mammalian mitochondrial reads 🙂 I am looking forward to connect with you 🙂
As a reminder: please feel free to invite anyone you think would benefit from the community here!
You do not have to have had attended one of the meetings in the past to join. Anyone and everyone is welcome (of course following the code of conduct)
Did someone attend the ISBA society meeting last week and could give a brief summary on what was discussed there? The only info I could find was the form that Matthew Collins shared on twitter (https://docs.google.com/forms/d/e/1FAIpQLSfM0T1HXAKVWCTIVMyDcSSufPRiR3FKTfoQmsauVF-gLTmjOg/viewform)
Anyone here use Krakenunique and can help me out? I'm trying to build a custom database
*Thread Reply:* Hi Abby, I can help you
*Thread Reply:* so basically, I have an fna file of reference genomes from a collaborator
*Thread Reply:* I've been trying to build a database (which is what I think I need to do)
*Thread Reply:* and that seems to work, except there are no classifications
*Thread Reply:* am I doing something completely wrong?
*Thread Reply:* @Ian Light as well I believe uses it
*Thread Reply:* @Abby Gancz yes, you build a custom database with krakenuniq-build from fasta file of concatenated reference genomes. I hope those are a number of reference genomes in your case, from multiple organisms, i.e. not just a few. By “no classifications” you mean that 100% of reads are unclassified?
*Thread Reply:* I am not sure whether I referenced the input file correctly though
*Thread Reply:* I made a 'library' folder and put the genomes in there, then run the build command
*Thread Reply:* Seems right. As far as I remember, krakenuniq creates a file_list.txt with the list of reference genomes included into the database, perhaps it is worth checking that all your ref genomes are there. Second, how heavy are your database.kdb files? I mean if something went wrong, they might be empty
*Thread Reply:* Is it microbial ref genomes or eukaryotic / mammalian?
*Thread Reply:* I would also add your ref genomes from collaborator to the other microbial genomes that can be downloaded by krakenuniq from ncbi. You need some “background genomes” for classification even though you expect most of your reads to be assigned to the ref genomes from collaborator
*Thread Reply:* Would a single fna file be fine as an input?
*Thread Reply:* I'm wondering whether that is the issue
*Thread Reply:* Abby, how many ref genomes are within the fna file?
*Thread Reply:* So it seems like the problem is in the database building step, where it's saying al the sequences are unclassified
*Thread Reply:* do you know how to deal with that?
*Thread Reply:* hmm, or maybe this is because I forgot the mapping file 😑
*Thread Reply:* This may be a dumb question, but is there a way to get taxonomy IDs from the fna file?
*Thread Reply:* hmm, the db building step says that the sequences are empty
*Thread Reply:* Abby, do you have headers in the fna-file?
*Thread Reply:* May I have a look at a few lines / characters of the fna-file?
*Thread Reply:* >NC_012975.1 Oryzias GCTCACGTAGCTTACCTAAAGCATGACACTGAAGATGTTAAGACAAACCCTAGAAAGTTT CGTAAGCACAAAAGTTTGGTCCTGACTTTACTGT
*Thread Reply:* Hmm, the ref genomes present in the fna-file should have a conversion assession2taxid-file. I think, it is not enough to just have a fna-file
*Thread Reply:* Hey @Nikolay Oskolkov, any chance I can get example files for KrakenUniq database construction? Including the taxonomy ID mapping
*Thread Reply:* @Abby Gancz did you figure this out? I am in a similar boat and maybe we can spare Nikolay from explaining everything!
*Thread Reply:* (specifically going from fasta to the mapping file)
*Thread Reply:* Still working on it, but I'd be happy to fill you in once we find a solution
*Thread Reply:* We do have a super clunky script that looks up the taxID and pastes it, if that helps
*Thread Reply:* Sigh I was afraid it’d be clunky script writing time… keep me posted!
@channel another reminder, if you have any ideas of projects/events/tools anything you think would be an interesting community/collaborative project, please feel free to add your ideas to the GitHub project board: https://github.com/orgs/SPAAM-community/projects/1
(If you don't have/want a github account, message me and I can add it for you)
:partyparrot:Hello @channel! @Ele and Shreya here, co-editors of the SPAAM blog!
📒 We wanted to tell you about what we have planned and invite you to join the <#C02D3DJP3MY|spaam-blog> channel if you are at all interested. We’re hoping this project serves as an open access resource to democratize the field, let us practice our science communication skills, and share practical advice and information.
🗓️ Starting in early 2022, we plan to post every month to 6 weeks. Posts will be one of two kinds:
🧠 Expert Explanations: Do you have opinions on a hot topic? Is there a concept you had to teach yourself because there isn’t a good explainer out there? Got something on your mind that you want to share less formally than in a journal? These will be 500-1000 word posts by a single author on a topic of their choice.
📧 Agony Aunt: Kraken2 or KrakenUniq? To UDG or not to UDG? You could spend forever reading threads on biostars… or you could take your question straight to the SPAAM community! We will solicit questions and pose them to SPAAMers to find out what people think (through polls/surveys and asking for more detail from those who are willing!). Then we’ll synthesize the various answers and write them up into a post!
✨ If you are at all interested in participating -- be it by suggesting a topic for either post type, writing an Expert Explanation, or being among the “survey participants” polled for Agony Aunt columns -- please do join the <#C02D3DJP3MY|spaam-blog> channel! And feel free to reach out to Ele or me with any questions or concerns :):sparkles:
*Thread Reply:* PS if you’re American we call Agony Aunts advice columns out here but I like the British version more ✌️:skintone5:
Dear all! I’m Guillermo, I recently joined SPAAM’s slack and also started immersing in the world of ancient metagenomics 🙂 I’m very excited about it!…. I recently started working with the nf-core/Eager pipeline and was wondering if someone here could help me with an issue I’m currently encountering?
*Thread Reply:* Hi @Guillermo Rangel and welcome! If the issue is specifically about nf-core/eager it would be better to ask on the nf-core slack under #eager (https://nf-core/join)
*Thread Reply:* But if your question is specifically to do with e.g. what parameter or database to use, for example, then you can ask on <#C02DCKJ54JX|no-stupid-questions>
*Thread Reply:* cool, thanks for your reply James 🙂 I’ll join both of those channels
Hello all!! I have one question for calculus-people: are there any study linking dietary elements (macronutrients?) to calculus bacterial communities? I've found some about oral microbiome (i.e., saliva) [ like these two just to give an idea: https://doi.org/10.1038/s41598-018-24207-3, DOI: 10.1111/mec.14435], while I am not finding anything about calculus in relation with these aspects, so I wonder if someone in the community has any information about that? I am actually working on some modern calculus samples and I found difference in genera abundance (16S) and found some correlation of that with carbohydrate, vegetable and alcohol consumption but unfortunately I have only general information about the diet of my samples (like consumption of cereals, meat, milk product etc. by week). Thus I wonder if someone has more stressed these aspects than me and maybe have more precise information like it has been done extensively for the gut. If not, maybe that can be an idea for a common project for this community?💡👀🙂
*Thread Reply:* Hi @Andrea quagliariello sorry for the slow reply, a lot of us were in an event for #ancientmetagenomedir!
AFAIK not much has been done for plaque/calculus in regards to diet.
Some papers have suggested there is a difference (Adler 2013, Weyrich 2017), but we were not able to replicate these results when we compared Gorillas/Chimps/HUmans etc (Fellows Yates 2021). However because of the complexity of the oral biofilm it might be quite hard to distinguish changes due to diet vs. other aspects.
In the SI of our paper (section S4.5) we also have examples of papers from modern microbiomes showing very very minor differences due to diet (see references in the last paragrph), and instead (our group anyway) think that looking at funtional differences will be a better way to track this, rather than taxonomic profiles.
*Thread Reply:* Hi @Andrea quagliariello there is very little work done on diet and calculus. To find papers, your best bet would be a PubMed search, order results by recent, go to the oldest papers, and move forward through them. Any work that was done is likely to be older, done in animals, and published in dental journals, ie https://pubmed.ncbi.nlm.nih.gov/5230026/
*Thread Reply:* Hello @James Fellows Yates and @ivelsko, thanks a lot for your reply. Don’t worry I knew you were all busy that day, unfortunately I was not able to join your meeting, hoping to be part of it next time. I took a look at pubmed and as I suspected there are very few information about the theme. @James Fellows Yates I totally agree with you; I think that the analysis of functional pathways may be more sensitive to detect possible dietary-linked influence rather than specific bacterial markers.
🚨 We will be running an #ancientmetagenomedir Metadatathon ROUND 2 in *December*! For anyone who couldn't make the first one, you are still welcome to join for this second event! I will run the same training sessions on Git(Hub) and AncientMetagenomeDir across all time zones as necessary!
PIs: you are also encouraged to join! It is good practise/skills building for you guys too (particularly given we are finding a lot of issues with how people have uploaded their data, so spreading awareness might be good 😉 )
Please fill in the Doodle poll to indicate the best time for you!
https://doodle.com/poll/v3aevnu5xi4w3ymf?utm_source=poll&utm_medium=link
*Thread Reply:* I'm going to shamelessly ping specifically 😉 @Katerina Guschanski @Nico Rascovan @Freddi Scheib@Felix Key @Hannes @Antonio Fernandez-Guerra @Pete Heintzman
If you wanna help make your student's lives easier downstream 😉 (also because in the future want to start talking about developing a standard MIxS template that ancient metagenomicists can use to upload stuff [currently without requesting new fields, but could extend in the future])
*Thread Reply:* Thanks, but unfortunately I can't take on anything new until after March 22. Maybe round 3.
*Aim*: aggregate and standardise library level metadata and make data accessible
*Objectives: • Training for use of Git(Hub) • Aggregate library level metadata for all raw sequencing data • Build a tool to allow efficient downloading of data based on metadata Result*: rapid filtering and downloading of pubilshed sequencing data. Any recorded contributor on the GitHub repostiory will be included on a paper presenting tools and discussion about issues about public data!
Oh and please share with your collegues/labs etc! The more the merrier
I finally uploaded my coverage/edit distance/deamination plotting script to github as promised during SPAAM3 so I am shamelessly plugging it here: https://github.com/MeriamGuellil/aDNA-BAMPlotter
*Thread Reply:* You should totally put it on bioconda
*Thread Reply:* Looking at the dependencies it would be quite straight forward
*Thread Reply:* haven't uploaded anything to bioconda yet (even though I love conda) I will check it out. Thanks!
*Thread Reply:* You basically just need to make one yaml file with download link of the release, md5sum, which license and then a list of dependencies.
*Thread Reply:* As in not even uploading anything
*Thread Reply:* Let me know if you want any help or advice doing it
*Thread Reply:* You even just copy Aida's endors.py file 🤔
*Thread Reply:* She's done it before
*Thread Reply:* Thanks @Meriam Guellil! Just tried it and it's awesome 😁
That also reminds me, while we are sharing, last week some of us released a github template on how we plan to standardise the structure of our bioinformatics projects in our group (where to place files, how to document things etc.): https://github.com/paleobiotechnology/analysis-project-structure
It's publicly available, and also works with the python 'cookiecutter' (thanks @Maxime Borry!) to insert your project name in all the relevant places.
Feedback welcome, and feel free to use it!
*Thread Reply:* Ooh just in time for me to try it out! 😁
@channel reminder of AncientMetagenomeDir Metadatathon round2 (*important*: you can still join if you DIDN'T join the first one!), please select a day that would work best for you here:
https://spaam-community.slack.com/archives/CPYE64ZC6/p1636362046018300
I will shut the doodle end of this week! So plesae sign up if yo uwant to be included in any future publication that may arise!
Also, if you ever want something (re)tweeted by the SPAAM community twitter account, please post your request on the #twitter channel!
Btw, if you've tried to signoff the SPAAM mailing list recently, but still kept getting emails about that. The wrong link was in the email footer, it should work now 🤦
@channel reminder of #events channel to keep up to date to ancient metagenomics relevant events/talks/workshops etc! I just posted details on a talk from @Maxime Borry about a pipeline for ancient paleofaeces today at 13:00 CET!
I've made a dedicated gather.town space for the SPAAM community 🙂
https://gather.town/app/PlXjb0deog0B4JCq/spaam-community
Feel free to use for any meetings/projects/socialising you wish to do within SPAAM! Let me know if you have problems accessing it!
Can anyone access the spaam website at the moment? It just gets stuck on loading for me 😞
*Thread Reply:* It loaded for me, but it took a moment
*Thread Reply:* Then it's just me
*Thread Reply:* OK now it works
*Thread Reply:* Thanks or checking!
SPAAM PROJECT NR. 3!!!
*I would like the announce the establishment, and call for members, of a working group for the creation of a MIxS checklist for ancient metagenomics and ancient DNA in general.*
MIxS checklists represent 'minimum information standards (checklists) – Minimum Information about any (x) Sequence (MIxS)'. These are essentially the standard set of metadata you are expected (required) report in publications. Domain-specific checklists already exist for metagenomes, genomes (MIMS/MIGS), marker genes (MIMARKS), metagenome-assembled genomes (MIMAG), uncultivated virus genomes (MIUViG), single amplified genomes (MISAG), parasite microbiomes (MIxS-PMP), argircultural microbiomes (MIxS-AG), biosynthetic gene clusters (MIBiG). They are maintained by the Genomic Standards Consortium.
These checklists affect your work as follows:
- These checklists are /mandatory/ for inclusion with upload of sequencing data to the SRA/ENA (i.e. you must fill them out)
- Are becoming more common to also be included in publications themselves, as SI
- They make your life better by making it easier to find useful public comparative data for your OWN research
We have have previously discussed such a thing in SPAAM2 for ancient metagenomics, but were recently contacted by researchers in ancient human population genomics, who were also aiming something similar for them. Therefore we are establishing working groups to work in collaboration to create a general checklist for ancient DNA sequences /in general/**. This is project is provisionally called MInAS (Minimum Information about aNy Ancient Sequence - thanks to Antonio Fernandez-Guerra)
We have also already been in contact with the ENA who are extremely interested in such a checklist. For further incentive, many of the MIxS extensions have been been published in visibile publications such as Nature Biotechnology and Nature Chemical Biology, showing the importance of such checklists.
**If you are interested and passionate about metadata, FAIR data, and/or reporting standards, and wish to be a part of the working group, please join the <#C01BX7EM4EL|metadata-standards> SPAAM slack channel, and indicate your availability on the following doodle poll for a first meeting: https://doodle.com/poll/d68p8zncarnvnkdt?utm_source=poll&utm_medium=link**
*Thread Reply:* Hi James, Is too late to join this interesting project? 🙂
*Thread Reply:* Nope! This got a bit delayed for a variety of reasons, I'm planning on setting up the next meeting tomorrow
*Thread Reply:* Great, thanks you James!
*Thread Reply:* @Mohamed Sarhan please join <#C01BX7EM4EL|metadata-standards> to keep up to date
Reminder for those who only recently joined the workspace - checked for the pinned message to see the most relevant channels for newcomers.
In particular the second paper coming out of SPAAM came out today - check #papers
Hi @channel! Happy New Year to everyone. @Meriam Guellil and I are doing a "small" project together in order to assess the choice of library construction + adapter trimming methods in ancient metagenomic analysis, with a focus in metagenomic classification of reads. For this we will need you help! I will create two polls to gaze which: which indexing/barcoding protocols are used in the field and which tools are used for removing adapters/barcodes/indexes. So it will be helpful to us if you could respond to the polls by the end of this month! Thank you all in advance!
Which indexing/barcoding protocol do I use? If adding an option, please provide a link to the paper describing the method. Also could you write in a comment the length of your indexes and/or barcodes
Hi all, I am very happy to join today the SPAAM slack thanks to Nicolas reminder. I created last year the sedaDNA scientific society - with many colleagues including @Pete Heintzman and it obviously took a lot of time to make it run but we have now a network of 300 members. There are shared interest between the members of our two communities and I am happy to serve as a bridge when it will be necessary. In terms of research, I am currently deep into ancient metagenomics analyzing metagenomes from Lake Ekoln (Sweden), the Black Sea and also trying to get my own Baltic metagenomes but it seems harder than getting 18S from sediments ^^. That is super cool that is some cases I can trace back functional genes from specific microbes. Also I got some MAGs from those sediments so I am interested about which one are low-energy adapted microorganisms that may have lived once in the water column. Hope to have opportunities to discuss with you all https://ercapo.wixsite.com/sedadna-society/ericcapo
Welcome! Really Great to have you! I think this is a good time as well for some starting since collaborations! We are just starting up a project to develop a MIxS checklist for ancient DNA, to improve metadata reporting. @Antonio Fernandez-Guerra and I had already talked about talking to the sedaDNA if they want to get involved, as we are already going to be working with people working in human population genetics. If you're interested, please join <#C01BX7EM4EL|metadata-standards>, we will be sending around a draft project proposal (for the GSC) next week to get more input.
We also have an ongoing project gathering and standardising sample and library metadata for all published ancient metagenomies (#ancientmetagenomedir), some more support for the sediment/environment papers would be great
Great thank James, happy to contribute where I can. I can also suggest certain members of the society that would be of good help for those projects. I am joining the two channels! Have all a good week!
Hi everyone, I am joining SPAAM thanks to the recommendation of Eric. I am working at the Senckenberg Biodiversity and Climate Research Centre in Frankfurt am Main, Germany (https://www.senckenberg.de/en/institutes/sbik-f/functional-environmental-genomics/), the Justus-Liebig University in Gießen, and at the LOEWE Centre for Translational Biodiversity Genomics (LOEWE-TBG, tbg.senckenberg.de). One of my current research interests is biodiversity time series generated with ancient sedimentary DNA (the other is functional environmental genomics of soil invertebrates). With sedaDNA I focus currently on Baltic Sea sediments and on lake sediments from north-western Germany. Happy to be here!
For everyone joining from the sedaDNA society, please see the pinned posts for possibly relevant channels: https://spaam-community.slack.com/archives/CPYE64ZC6/p1632471482043200
I’m currently in the process of putting our (UTIG) protocols on protocols.io and I have some questions/would like to have some exchange with someone more experienced. Since I know at least a few people from Jena/Leipzig used it before and probably more people are interested I was wondering if it would be worth creating a channel for protocols.io specifically. What do people think about that?
*Thread Reply:* Maybe #wet-lab would be more suitable?
*Thread Reply:* (to make it more broad?)
*Thread Reply:* or just #protcols
*Thread Reply:* But agree in general thereisn't a really suitable place atm
Actually this is a good question for @channel - when joining a new slack workspace, how do you normally find relevant channels you want to join?
(new channel is called: #wetlab_protocols 🙃)
Thakns all! I won't do anything fancy with the pinned messages then 😄
I do not know if it is a right place to write it here but I just wanted to express my gratitude to @James Fellows Yates for his wonderful rma-tabuliser https://github.com/jfy133/rma-tabuliser that I really recommend to everyone. Very easy to use and it gives me exactly what I want. I remember I said previously that rma-tabuliser seems to duplicate MaltExtract output but I can see now that rma-tabuliser is much more comfortable so I just want to thank James here! 🙂
*Thread Reply:* You're welcome :)
Do check the output though, I never tested it super thoroughly...
*Thread Reply:* As in I nicked the column merging awk command off of stack overflow
*Thread Reply:* And it worked in all my tests but I still don't really understand how it does it 🤣😬
*Thread Reply:* https://github.com/jfy133/rma-tabuliser/blob/main/rma-tabuliser#L267-L271
*Thread Reply:* Ha-ha, thanks for pointing out! 🙂 Quite a piece of code 🙂
@channel I've just made live the new SPAAM website (inspired by the sedaDNA's website - thanks @Eric Capo 😉 )! Please have a poke around and let me know if you come across any issues.
This is in preparation for the upcoming <#C02D3DJP3MY|spaam-blog>, so keep 👀 open!
*Thread Reply:* First issue is that the logo on the left corner is cut. That depends on the size of your screen. Try on other screens and you will see. The way to overcome is to have shorted text below (or less links)
*Thread Reply:* yeah @Maxime Borry also noticed a problem on mobile too
If you want the nice bubbles popping like in our website, feel free to fork us (and modify background colors, direction and shape of bubbles).
*Thread Reply:* I was actually trying to do that, but I was trying by forking the original hydrout and copying the bits I wanted from sedaDNA
*Thread Reply:* Didn't work so well it seems 🤦♀️:skintone3:
*Thread Reply:* yes I struggle hard too. So my way was to fork from mersorcium and play a bit with parameters later.
*Thread Reply:* But it indeed took me few days to get how to do that ^^
*Thread Reply:* If you fork directly the sedaDNA website, I could guide you to change colors and shapes
*Thread Reply:* 👍 I have a closed-Kita situation atm, but I'll look into it when I have free time again! Unless you have time to make a fork from yours in the SPAAM organisation and tweak it as per what is already there? Regardless, I will add you ot hte SPAAM github
*Thread Reply:* you should have an invite
*Thread Reply:* Overloaded right now too, sorry but I will later
Reminder: the SPAAM gather.town is completely open to everyone in spaam for any purpose!
@channel there are some interesting #jobs adverts on... well #jobs , please share in your networks too!
A word of gratitude again 🙂 Have just tried PyDamage on my assembled contigs, and I am a fan of PyDamage, thank you very much @Maxime Borry 🙂 Really recommend it to everyone who does de-novo assembly of ancient microbes. I previously ran mapDamage (with --no-stats flag of course) and PMDtools on each contig, and did a simple Mann-Whittney U test on AUC for transition frequency against all other mutations, but am going to switch to PyDamage as it seems to be more efficient than my approach. @Maxime Borry is a 🌟
Hi @channel,
We are very excited to announce that SPAAM4 is currently in the making and will be held virtually in Autumn 2022 🎉
Similarly to past years, we have designed a questionnaire to help us tailor the conference to the SPAAM community, to gauge interest and ensure we can all get as much out of this conference as possible!
Please fill in this questionnaire by the Friday 25th March https://forms.gle/eLqN28AxKUHCZJfe9
Also remember to follow the SPAAM twitter account, slack channel and join the mailing list (which can all be found at tinyurl.com/snj9azch) to ensure you don’t miss out on SPAAM4 updates!
Helping all your ancient metagenomic dreams come true,
SPAAM4 Organisers @Abby Gancz @Gunnar Neumann @Nasreen Broomand @Rita M Austin @Shreya @Pooja Swali
(Please feel free to contact any of us with any further questions!)
@channel we are arranging our (hopefully) final library metadatathon for AncientMetagenomeDir. Once all the metadata is in, we can finalise a new downloading tool for ancient metagenomic data, and are planning on publishing this. If anyone is interested in joining in - please register your interest on the doodle on #ancientmetagenomedir ! We will run training on GitHub and AncientMetagenomeDir, so newcomers are welcome 🙂. Deadline is end of this week!
Congrats to @Hannes on your erc consolidator!!
*Thread Reply:* Tienes una problema... @Nico Rascovan
*Thread Reply:* hahaha, with the nicknames ?
*Thread Reply:* I only use them with the people that I like 😉
*Thread Reply:* well that's just wierd
*Thread Reply:* The diminuitives yes...
*Thread Reply:* it’s even worst, not only diminutives, but I can come with any sort of weird name… but they are all stemming from pure love.
Hello @channel!
Just a short reminder: Please, remember to fill out our questionnaire for this year’s SPAAM4 meeting by Friday 25th March. You have three days left. https://forms.gle/eLqN28AxKUHCZJfe9
Your input will help us to find a date that will fit as many people as possible in our community and to prepare a program to spark the interest of everyone!
Congrats to @Ele!!!
*Thread Reply:* It was a perfect poosentation!
Hi @channel!
I just want to give you all, as exclusive members of SPAAM, a pre-notification of something that is going to be announced on Friday:
I'm pleased to announce that we will be running our first ever SPAAM Summer School this summer, with this particular edition being called 'Introduction to Metagenomics'. It will be primarily aimed at M.Sc., and early Ph.D. students, and will be a 5 day course to give students a guided practical hands-on run through of data analysis, from how to use the command line to performed De Novo assembly and phylogenomics.
It is open to a maximum of 25 people, and will be publicly advertised. So please keep your eyes open on Friday for the various announcements for more information, if you're interested!
Cool! Will it be online or in person? Or do I have to wait until Friday to find out? 🙂
Online (with hybridish if we have people from nearby). But all other details come on Friday 😉
So amazing!!!! What a great initiative!
*Thread Reply:* Yay for non-standard funding!
*Thread Reply:* Will you disclose your sources?
*Thread Reply:* The Palaeobiotechnolgoy one from the Werner Siemens-Stiftung: https://www.wernersiemens-stiftung.ch/en/projects/ancient-medicine
*Thread Reply:* Tina and Pierre also added a request for teaching/training support too
*Thread Reply:* Great stuff!
omg I want to come! Was planning a protein summer school, but may have been beaten by this! ha
@channel summer school is now official announced! https://twitter.com/spaam_community/status/1519973404866224130

Registration is now open 🙂 (I hope)
*Thread Reply:* Any chance of auditing (or will there be recordings)? My summer schedule is a total mess and I also don't think I qualify as early PhD 😛
*Thread Reply:* We plan to make everything available online after, but no guaruntee
Please share amongst your colleague/networks too!
*Thread Reply:* @Eric Capo maybe you oculd share with the sedaDNA mailinglist/twitter?
*Thread Reply:* Yes I will definitely do that (within next week). Well done for organizing that!
*Thread Reply:* @James Fellows Yates Maybe adding the word "sedimentary DNA" or molecular data preserved in environmental archives to the description of the summer school description would help you to get some people from the society. Because the description now is about "degraded DNA content of archaeological and paleontological specimens" and that excluded "molecular ecologists"
*Thread Reply:* That's actually on purpose 😬
*Thread Reply:* We will focus almost exclusively on host-associated stuff, because that is where our expertise is. However may of the concepts are transferable, as we will be going quite basic like - how to work on a terminal, and this is what a taxonomic profile does - rather than 'how to pick database, or select X markers for Y context).
*Thread Reply:* Ok I understand that this need to be specific so but then I was confused by this sentence in the description of the summer school "how past ecosystems changed in response to long-term climatic and anthropogenic change". This is clearly targeting environmental archives right?
*Thread Reply:* I have a Romanian colleague who is technically not a student right now. She will be starting her master's in September. Is it okay for her to apply?
*Thread Reply:* Great thank you!
Hi all! It's the moment you've all been waiting for--SPAAM4 dates are now announced! Mark your calendars, SPAAM4 will take place October 12-14, 2022. That's 3 half-day sessions full of science, microbes, and maybe...romance? A lot of love for microbes, at least! There will be conference-style presentations, community-building opportunities, and more. At SPAAM4, the world is your oyster (unless you're Vibrio vulnificus--then maybe the oyster is your world?). Registration and abstract submission forms will be posted in the next few weeks. SPAAM4: The perfect marriage of conference and community. Don't miss it!
@channel reminder that the application form for the SPAAM/Werner Siemens-Stiftung Intro to Ancient Metagenomics Summer School is shutting in just over a week! Info here: https://spaam-community.github.io/wss-summer-school/
Also, as a side note, anyone else in the community is welcome to develop other summer schools/training courses (more advanced, or more specific topics) under the SPAAM 'name' as long as the topic is relevant to ancient metagenomics! Just let me know and I can help get you set up with the website system :)
*Thread Reply:* Really looking forward to this! I remember reading somewhere that in-person participation may be possible. Is there any news?
*Thread Reply:* We are waiting to see who we select (currently have 3:1 applicant: place ratio) and will make a decision then
@channel Registration and abstract submission for SPAAM4 are open! In case you missed it when we sent out our beautiful and very normal Save The Date: SPAAM4 will happen over three half-days (October 12-14, 2022). There will be conference-style presentations, community building opportunities and more. Spots are limited, so snag your spot early!
Register here: https://tinyurl.com/SPAAM4Reg Submit an abstract here: https://tinyurl.com/SPAAM4Abs
@channel we also created an <#C03HR4RLTGV|spaam4-open> channel for any questions, suggestions, ideas regarding SPAAM4. Come join!
@channel I've just discovered Slack has rolled out for all plans the ability to display 'Pronouns' (and soon name-pronunciantion guides!) on Slack profiles. I've now activated this for the SPAAM workspace.
To update:
- Click your face
- Press 'Profile'
- Press 'Edit profile'
- Add your pronouns to the field
- Press Save changes. then if you click on someones name, or press your own face > profile again, you should see them under your name.
I highly recommend adding pronouns and also pronunication (when htat's rolled out), particularly as we start expanding outside western/European-baesd languages to make it easier for everyone to communicate and chat with each other :)
@channel for all the recent newcomers, usful announcement channels to join:
#general #jobs #papers #events <#C02DCKJ54JX|no-stupid-questions>
And of course explore all the other channels that may be relevant for you 🙂
Hey all, has anyone used the OAGR database in the past and is having trouble downloading data from it now (security/access issues)? Wondering if this is a temporary issue or if the site is no longer maintained
*Thread Reply:* I remember this happening to @James Fellows Yates who figured out that they changed the url
https://oagr.org/ (they dropped .au) and if you're looking for Weyrich 2017 data, it's since been uploaded to the SRA :). You can check AncientMetagenomeDir
@James Fellows Yates Exactly what I was looking for, thanks!
*Thread Reply:* If you run into any issues, I should be able to help. I am the one who uploaded them.
*Thread Reply:* Downloaded without any issues, thanks Sterling!
As Shreya, not as a SPAAM4 organizer or anything, I'm having a hard time thinking about microbes today. For my US people, just wanted to mention that you all know someone (me) in Illinois. In case you or someone you know wants a trip to a state like Illinois for whatever reason, I'll always be happy to host you. Okay, back to the microbes, and sorry to get off topic. Register for SPAAM4!
Dear @channel ! Just a little reminder to register for the upcoming SPAAM4 meeting in October! Register here: https://tinyurl.com/SPAAM4Reg Registration deadline is September 12th.
Also, share your exciting work, results and thoughts with the SPAAM community and submit your abstract here: https://tinyurl.com/SPAAM4Abs You have less than a month left as submission deadline is August 1st!
Congrats @Pete Heintzman!

Thanks, @James Fellows Yates and everyone! 😃
Hello! Did anyone here work on this paper and could help me track down Supplementary Table 2? Can’t seem to find it in the PDF. Comparison of target enrichment strategies for ancient pathogen DNA, Furtwängler et al 2020: https://www.future-science.com/doi/full/10.2144/btn-2020-0100
*Thread Reply:* Oh wow... Table 2 and 3 are missing 👀
*Thread Reply:* Anja is in our department, I'll ask her
*Thread Reply:* Apparently the publisher didn't upload those tables... They were in a separate file. Anja sent them to me (attached) and said she'll ask them publisher to upload them as well :)
*Thread Reply:* OMG thank you James!!!!
Hi @channel! Bummed that you missed the SPAAM4 abstract submission deadline? You're in luck! We are extending the abstract submission deadline for SPAAM4 to August 15. The abstract doesn't have to be fancy or discuss results, just pitch us your ideas! Submit here: https://tinyurl.com/SPAAM4Abs
@channel SPAAM is now an affiliate organisation to the larger ISBA society (international society for biomolecular archaeology) !
We will likely be receiving support from ISBA for future workshops/events etc., so please do follow it's various social media(s) to keep up-to-date! Furthermore the next ISBA Symposium meeting will be happening in Tartu 2023, and there will likely be LOADS of ancient metagenomics there - so keep informed!
The (pre-)launch message from the provisional secretary (Oliver Craig, UoY)
```It is a pleasure to (pre-)launch our new International Society for Biomolecular Archaeology (ISBA) and announce the date for the next meeting, our 10th!, which will be in Tartu, Estonia on the 13-16 September 2023.
Details of the society and the meeting can be found on our new website here: https://www.isbarch.org/
We are still waiting for the outcome of our application to the UK charity commission to register our society. Hopefully once this is in order, you will be able to apply for membership by following the link on the new site.
You are receiving this message as you gave consent during the last ISBA meeting to be contacted but do feel free to publicise more widely through your channels.
For future messages and updates - please join our official mailing list: http://www.jiscmail.ac.uk/ISBARCH
You can also follow us on twitter @ISBArchaeology
Thanks to everyone who has worked so hard behind the scenes to make this happen! ```
@Maxime Borry and myself (among others) are looking for some help/suggesitons:
https://mobile.twitter.com/me_beber/status/1563926235717173248
What were you thinking? Were you thinking of something similar to the Velosko et al 2019 paper?
No, Moritz explains later in the thread that we are building a pipeline that will do taxonomic profiling across multiple tools and databases in a highly parallelised manner, because as we all know (and Irina shows), different tools/databases give different results.
But at the same time, different tools have output formats. So we are building a tool that standardised these for easier comparison. But then Mortiz and Maxime were curious if there would be a way to make a 'consensus' across the different tools/databases, to see how well they agree. But we quickly realised this won't be trivial, thus the question for input
*Thread Reply:* I see now. I would be interested in knowing how this could be done without taking an exorbitant amounts of memory. We run into this issue a lot with our current workflow.
That said, with the pipeline you could indeed do a repeat of Velsko 2019 quite easily
*Thread Reply:* I don't think you can assume that mineralization will destroy/degrade the DNA. Hydroxyapatite binds DNA very well, and this actually protects it from breaking down. B/c of that property, hydroxyapatite-DNA binding is used for industrial purposes
*Thread Reply:* Do we know how they are protected from degeneration by HA? since we don't entirely know the exact mechanism of mineralisation, could it cause damage to the DNA (even before binding), if, as proposed in an an article I read but can't quite remember, the mineralisation of bacteria starts from within the cell?
*Thread Reply:* The negatively charged backbone of DNA binds the positively charged HA molecules, which then protect it from degradation by hydrolysis and enzymatic activity. I've heard the hypothesis that mineralization is nucleated by the DNA, and the HA molecules grow up the backbone
*Thread Reply:* If you google "hydroxyapatite growth on DNA" you'll get a lot of papers about hydroxyapatite growth on DNA. All synthetic systems, but they probably describe how it happens, and it's possible the same occurs in bacterial cells
Shame(ful) self promotion: https://twitter.com/jfy133/status/1567051387426439171
If anyone here is using eager, please let us know what you're unhappy about/what improved!
@channel Who here is working on ancient microbiomes and would like to review a journal article?
I agreed to review a paper, but they are looking for suggestions for a second reviewer (don't be shy even if you're early in your program - it's a good to practise !)
*Thread Reply:* If no one else has showed interest, I can do it.
*Thread Reply:* I'll add you to the list
*Thread Reply:* If suggestions
*Thread Reply:* That sounds great! Thank you
If you're interested let me know and I can suggest you
(and it's not at a super high prestiguous journal and you can remain anonymous, the journal has a nice interface too 🙂 )
What do does everyone call the metric, of the number of reads that align to your reference genome?
e.g. endogenous DNA, or percent-on-target etc. do any of you have any other terms?
*Thread Reply:* I think endogenous is most common but also imprecise. If I could choose I would go for “target”. There might be cases where you actually want to look for an organism that’s “exogenous”.
*Thread Reply:* Yup, I agree. But I'm looking for a list of all possible terms 😬
*Thread Reply:* yes I stopped using endogenous because it made little sense for pathogens. I usually go with something in the lines of %target taxon/refseq
*Thread Reply:* refseq being?
*Thread Reply:* depends on what I was using but chances are when you map you don't have a pangenome reference so taxon can be misleading too
*Thread Reply:* I would also use “XX targeting reads” (on a given genome)
*Thread Reply:* I meant more specifically the normalised version that allows you to compare (and set a threshold for Y/N on sufficient preservation)
Is there anyone here who is using KrakenUniq and happens to know:
- An official tool to merge output of multiple samples into one table
- Whether you can run multiple samples for one execution of the tool? (rather than having to have one command == one sample)
*Thread Reply:* https://github.com/jenniferlu717/KrakenTools
*Thread Reply:* Thats Kraken only
*Thread Reply:* Or rather whne I ran the command
*Thread Reply:* it gave an error
*Thread Reply:* But it does say in the description!?
*Thread Reply:* Since they state "KrakenTools is a suite of scripts to be used for post-analysis of Kraken/KrakenUniq/Kraken2/Bracken results." you'll have to email them
*Thread Reply:* I think outside of these tools there is only pavian as an "official tool"
*Thread Reply:* thakn you both!
*Thread Reply:* @James Fellows Yates regarding your first question, I am using a custom R script that combines multiple KrakenUniq outputs into a single abundance table, can share the script if you want, it is also included into our upcoming workflow release. Regarding the second question, I unfortunately use one command line per sample, would love to know if it is possible to use multiple inputs. I got a feeling from @Meriam Guellil’s presentation last SPAAM that she loaded the DB only once and processed multiple fastqs. This is probably what you are looking for, right? If so, I never managed to do it like Meriam but perhaps I am not aware of some magic flags 🙂
*Thread Reply:* I think krakentools does support combining report-style output apparently, but I'm getting an error I'm looking in to
*Thread Reply:* Thanks for the offer on the script! I'm looking more for an official tool (basically already on bioconda), but I might come back to you for that
*Thread Reply:* yes I load the DB once and then loop through the files. And I combine the outputs in python using mostly pandas
*Thread Reply:* Do you have a little code snippet you could share @Meriam Guellil?
*Thread Reply:* or rather db load and loop
*Thread Reply:* This should do the trick:
krakenuniq --db ${DB} --preload --threads ${threads}; while read sample; do
krakenuniq --report-file REPORT --fastq-input FASTQ --gzip-compressed --db ${DB} --threads ${threads} --only-classified-output > KRAKEN
done < sample.list
*Thread Reply:* with some variable shenanigans for the file names
*Thread Reply:* thank you! that's really helpful!
*Thread Reply:* I've used Pavian to do this is the past, although it's not an ideal solution.
https://github.com/fbreitwieser/pavian
*Thread Reply:* Yeah I need a cli tool unfortunately
*Thread Reply:* Thanks @Meriam Guellil for sharing! I am just curious, wasn't the --preload thing broken until recently?
*Thread Reply:* always worked for me but I think it was only broken in one particular version and now they switched it to implement the new loading system (I haven't changed to that yet) but 🤷♀️:skintone2:
*Thread Reply:* according to their github it was fixed for v0.6 in October 2021 but not quite sure which version broke it. I install it through conda and I guess I missed it somehow
*Thread Reply:* As far as I know many people from different groups tried --preload and reported that it was not working but I suspect it might have something to do with your particular cluster environment
*Thread Reply:* either way it should be working now
*Thread Reply:* @Nikolay Oskolkov I have been using the --preload option on Kebnekaise and it works fine. Also on Kebnekaise we could preload the database into a cluster ramdisk making it possible to only use the same preloaded database for all the runs in the same slurm array of jobs. Gave the same results than without the --preload option on Uppmax. Nevertheless, I believe this trick is quite cluster specific. (Sorry guys those are just weird Swedish cluster names)
*Thread Reply:* I've heard of uppmax but Kebnekaise is a new one😅🤣 how do you even day that?!
*Thread Reply:* I just try to say it and probably fail every time, Swedes won't complain anyway 😅 Sadly that cluster won't be available at the end of December, I was happy there 🥹
Hi everyone! If you are attending SPAAM4:
- join the <#C03HR4RLTGV|spaam4-open> channel before the conference!
- We are looking for 3-4 people to be SPAAM4 Code of Conduct Officers. Hopefully this is a very boring job! It would entail having your name on the SPAAM4 program as someone that attendees can come to in confidence if they need to report a Code of Conduct violation. You could then flag the issue to us organizers so we can deal with it. Ideally the CoC officer set represent some diverse languages/genders so people feel comfortable approaching them, and does not completely overlap with the organizers group! Please comment or DM me if you’re interested. Thank you!
*Thread Reply:* @Betsy Nelson is that a yes? We would love to have you on board again 🙂
Is there anyone here from a non German or UK country who wants to sign up as an ISBA member and wants to help troubleshoot something 👀 ?
*Thread Reply:* I haven't signed up yet, if the Netherlands would help?
*Thread Reply:* Do you have a spare 5 imnutes now?
*Thread Reply:* Or in the next 15?
*Thread Reply:* https://gather.town/app/PlXjb0deog0B4JCq/spaam-community
*Thread Reply:* I just signed up as a member last week (I’m US-based). Let me know if there is anything I can do to help!
*Thread Reply:* Thanks! Had a few people help me out, unfortunately for many countries paypal is a :poop_party:
*Thread Reply:* So we are looking for alternative solutions too now
Dear all, we are making a few large KrakenUniq databases and Bowtie2 indexes public:
```KrakenUniq database based on full NCBI NT: https://doi.org/10.17044/scilifelab.20205504
KrakenUniq database based on microbial part of NCBI NT: https://doi.org/10.17044/scilifelab.20518251
KrakenUniq database based on microbial part of NCBI RefSeq: https://doi.org/10.17044/scilifelab.21299541
Bowtie2 index for full NCBI NT database: https://doi.org/10.17044/scilifelab.21070063
Bowtie2 index for pathogenic microbial species of NCBI NT: https://doi.org/10.17044/scilifelab.21185887``` As you may know the new version of KrakenUniq allows running taxonomic classification in low-memory computer environments (in theory, even on a laptop) provided that a database is available. Further, Bowtie2 aligner can be used for following up (authentication, validation) KrakenUniq results. Bowtie2 is fast and quite memory-efficient, and one can use e.g. sam2lca from Maxime Borry and co https://www.theoj.org/joss-papers/joss.04360/10.21105.joss.04360.pdf to add an LCA on the top of Bowtie2 alignments. You can download the KrakenUniq databases and Bowtie2 indexes and use them for you research, we only kindly ask you to cite our work here https://www.biorxiv.org/content/10.1101/2022.10.03.510579v1. Please contact me if you discover any problems downloading or using the databases.

*Thread Reply:* Just a short comment. The NCBI NT is the one used by default by BLASTN. The Microbial NCBI NT includes: archaea, bacteria, viral, fungi, protozoa and parasitic worms (plus it also includes hg38 human references genome + a few complete eukaryotic reference genomes to be served as a decoy to attract potential eukaryotic contamination)
*Thread Reply:* Hey @Nikolay Oskolkov, I just wanted to clarify if the Microbial NCBI NT database is from all microbial genomes as per June 2020 or 2022? On figshare it says 2020 in the title but 2022 in the text. Also, thank you so much for making this available!!
*Thread Reply:* Hi @Meriam van Os the microbial NCBI NT if from June 2020. I have just checked the README.txt and it also correctly says "June 2020". Where did you see 2022?
*Thread Reply:* oh @Meriam van Os I have just found it! Indeed, in the short description for that database there was 2022 instead of 2020, sorry about this, I have just fixed it, and it should be updated soon. Anyway, it is the 2020!
*Thread Reply:* I was just trying to find in on my phone haha. Thanks for your quick reply Nikolay! I'll give it a try soon 😁
Hi @channel! SPAAM4 is approaching in just a few days! Friday will include a session on the “Future of SPAAM” and to facilitate discussion and aspects that you as the SPAAM community want to talk about, James has already prepared a google doc where everybody can already put in their thoughts and ideas. This can be anonymous, but indicating your name might help to follow up in case sth might be unclear. You can find the doc here: https://docs.google.com/document/d/1a6opWD70K9NAV0C3uo6ZVZg6VGjCbFxPXOsGS5N8HfE/edit?usp=sharing
also, SPAAM4 attendess, dont forget to join the <#C03HR4RLTGV|spaam4-open> channel!
Hi all :) Do you know of any tools/pipelines used to remove contaminations from ancient metagenomic data? Ex: DeconSeq Rather than being interested in tools for contamination assessment or quantification, I am interested in the removal of the contaminant reads from the fasta/fastq files. Thanks!
*Thread Reply:* I believe recentrifuge is supposed to be able to do this
*Thread Reply:* The Decontam R package wouldn’t necessarily remove reads per se. It would remove mapped reads to contaminants which may be helpful in some cases if you had cross contamination and your sample reads infiltrated your EBCs.
*Thread Reply:* Right, and for decontam you need additionally either [DNA] or negative controls.
Question for all MALT users: which MALT version are you using? I tested v0.5.2 last year and had a lot of issues with it. The biggest issue was that v0.5.2 didn’t build the database properly. For example, I supplied pathways to three different directories containing different subsets of references and it only included the first directory/pathway supplied. I tried many different ways of supplying the references and tested it by running samples where I knew what to expect to find. This is very different to the other version I’ve been using v0.4.1. In the end I gave up with v0.5.2 and I decided to stick with v0.4.1. Am curious to know what are other peoples’ experiences are with the newer MALT version(s)?
*Thread Reply:* Interesting. We have been using v0.5.2 but haven't been running into any issues. Just so I understand, you are having trouble with malt-build and malt-run?
*Thread Reply:* @Åshild (Ash) we have also discovered lots of issues with the recent versions and therefore have been using 0.4.1 as the most stable version to our experience. However, this version also has bugs, a colleague of mine, Claudio Mirabello, has reported an issue here http://megan.cs.uni-tuebingen.de/t/malt-build-does-not-seem-assign-a-tax-id-to-some-references/1758
*Thread Reply:* @Sterling Wright Yes, I had issues with malt-build (it wasn’t functioning like v0.3.8 or v0.4.1), and I already knew about the LCA issue that Aida pointed out as well.
*Thread Reply:* @Nikolay Oskolkov Thanks for sharing. I didn’t know about the taxon ID issue
we have also been doing some quite extensive testing and realised that the LCA algorithm is not properly working. Of the versions that we tested v0.3.8 seem to the one that still performed as expected. Version 0.4.1 apparently has some issues to assigning the reads to species that are highly represented in your database. I have decided also to stick to v0.4.1 too. Arthur left a few issues in the MEGAN comminunity:
for the version 0.4.1 issue: http://megan.informatik.uni-tuebingen.de/t/unable-to-change-the-default-coverage-parameter-for-naive-lca-assignment-with-malt-v-0-4-1/2032
and for the issues with v0.5.2 and v0.5.3 : http://megan.informatik.uni-tuebingen.de/t/lca-placement-failure-with-malt-v-0-5-2-and-0-5-3/1996
Thanks Aida! Glad I’m not the only one who had issues with it. Hopefully the developers will fix it
I got a response a couple of weeks ago from Daniel saying he has been very over capacity recently, but he would try and look soon. I'll try and nudge him again
general wetlab protocol's question: what is your favorite extraction protocol for dental calculus?🤔
*Thread Reply:* https://www.protocols.io/view/ancient-dna-extraction-from-dental-calculus-bidyka7w
@channel <#C02D3DJP3MY|spaam-blog> is our latest community project, to use @Nasreen Broomand phrase, you can think of this like spaam wikipedia. We would like to grow the blog as a really useful resource for people in this community. It'll be a great place to learn and to write a fun piece that will help future researchers. We'd love you to write us an article 😄 You can see more info here: https://spaam-community.github.io/blog/2022/07/15/blog-welcome/
*Thread Reply:* @Ele and @Nasreen Broomand: I don't know if you got that in the zoom chat, but I'd love to help with this in a couple of months
*Thread Reply:* Lovely!!! We will be in touch Maria!
Hello @channel, to follow up on the future of SPAAM discussion, please weigh in on the above: should we stay here on Slack or move to a different platform? Feel free to add other options and discuss the pros and cons in a thread here!
*Thread Reply:* It’d be great to have a platform that doesn’t erase messages, but I honestly hate that Discord doesn’t allow you to select which channels to be in 😑It means you get notifications for everyyyyy channel (you can mute but the highlighting for new messages never goes away since you cant actually leave it). I’ve never used Element, but the channels thing is what keeps me attached to Slack. Also replies don’t thread 😑
*Thread Reply:* Slack is the obvious choice for me too, the only downside is the disappearing messages. I’ve lost a couple messages where people gave me help that I wanted to keep referencing… I guess as it grows we could have a pinned post telling people how to use the slack, including “if someone helps you, save the message to your own notes”
*Thread Reply:* I tried Element a while back and it really doesn't compare to slack (of course that was before the 90-day message BS...). Might be worth piloting a few options to see what works; and worst case just stick with slack
*Thread Reply:* How many members are we currently? It looks like if we upgrade to Pro you will get back your messages from >90 days ago (which are only hidden, not deleted)
*Thread Reply:* That's the current stats
*Thread Reply:* Also just found out that Element is beta-testing replies in threads!!
*Thread Reply:* But do they support gifs?
*Thread Reply:* It looks like they have a giphy integration
*Thread Reply:* I think only if you host your own server though 😕
*Thread Reply:* From the one I created for the Rchaeology group (not self-hosted)
*Thread Reply:* @Olivia It's possible to select which channel you wish to receive notifications for in Discord -- right click on the channel and select 'Mute Channel' or 'Notification Settings'
*Thread Reply:* Not being able to see messages past 90 days is a massive hindrance to sharing information/knowledge IMO. Discord is everything that Slack is + more, and it's free.
*Thread Reply:* @Raphael Eisenhofer that's a fair point, althoug hthe entire opt-in by default is a bit shit (IMO), I'm also concerned that discord could also start locking down in the future (and we would have to go through the whole hoopla again))
*Thread Reply:* Which is making me leaning towards just trying to get some funding anyway...
And one more @channel please remember to review AncientMetagenomicLabs!
https://spaam-community.github.io/ancient-metagenomics-labs/
Plesae check your lab is listed and all the information is up-to-date. We hope this will be a useful resource for finding collaborators, reviewers, job opporunities etc.!
@channel I'm happy to announce our next community event!
We are calling anyone familiar with python 🐍 , or who wish to be a guinea 🐷 for wrapping up development and testing of our #ancientmetagenomedir ToolKit tool (AMDiRT), to get the tool and dataset ready for publication!
We will be having a two day hackthon Weds 9th-Thurs 10th November from 09:00_16:00ish CET (and possibly longer for other timezones, depending on attendance)! Please add this to your calendars!
@Maxime Borry will be leading the development team. Also, if you're not comfortable with python or software testing, you are also welcome to use the time to help the community 🤝 get in more publications into the #ancientmetagenomedir sample and library metadata database, as I will be around to help out with this 🙂.
Anyone who contributes either to the data and/or the tool will be included as coauthors on an upcoming publication 📑.
If you're interested in helping out please join #ancientmetagenomedir
And it will be on the SPAAM gather.town!
So remote attednecne possible 👍
For the people who asking about MALT a couple of weeks ago - apparently two days after he said he would work on it he was very sick for two weeks, he's now recovered so he will try and look again ASAP
http://software-ab.cs.uni-tuebingen.de/download/malt/welcome.html
v6.0
Ok bad news: @Arthur Kocher found a couple of other bugs (but aren't as deal breaking), but Daniel is aware
Good news, already fixed! http://software-ab.cs.uni-tuebingen.de/download/malt/release_notes.txt
Dear @channel for those of you interested in community informal discussions! We have a new channel #spaamtisch In this channel we will socialise and talk about community decided topics. Please join the channel if you are interested and: 1. Vote for a day that works for you to have the first SPAAMTisch chat 2. Vote for a topic among the listed ones. Thanks!!
Who else here is on Mastodon? And if so do you want to swap handles (post in thread :) )
*Thread Reply:* I'm: @jfy133@genomic.social
*Thread Reply:* @James Fellows Yates could you please explain me why people are moving to Mastodon? What is wrong with Twitter?
*Thread Reply:* Elon Musk has fired half the employees, of the remainder half (or more) have quit. Also he plans to monetise it like crazy to make up the money he's all lost, and he's essentially allowing more abuse and shit on it in the name of 'free speech'
*Thread Reply:* Aha, ok, I am not up to date on this topic 🙂 thank you for the explanation!
*Thread Reply:* A quick skim here also explains why people are moving to mastodon: https://www.washingtonpost.com/politics/2022/11/12/musk-is-wrecking-speech-moderation-twitter-theres-an-alternative/
*Thread Reply:* (basically better networking on specific topics, better moderation)
*Thread Reply:* The timeline here also describes worrying behaviour: https://www.bbc.com/news/technology-63675849
*Thread Reply:* Such as the banning of people joking about musk
*Thread Reply:* Free Speech! (just not about me...)
*Thread Reply:* @Gunnar Neumann@ecoevo.social
*Thread Reply:* I haven’t joined Mastodon yet, but will soon! I checked Twitter a couple times in the past two days and I experienced some performance issues already, like replies to tweets not loading etc.(even though my internet is fine)
*Thread Reply:* @zoepochon@genomic.social
*Thread Reply:* sorry guys I am a bit behind on this topic. I would also like to join but haven’t done a profile yet. I will soon!
*Thread Reply:* For those who are joining/already joined don't forget ot add yourself here: https://jfy133.github.io/Mastodon-Palaeogenomicists
Hey! Not metagenomics, but I was wondering if anyone has ever come across the following. I've been capturing ancient samples for mitogenomes, and in my latest batch, the libraries seem to be 100bp too long (~260bp on the BioAnalyzer) compared to other batches I've done with the same protocol. I doesn't seem to be "bubble peaks" from PCR overamplification as we tried denaturing them, and the peak stayed the same. Has anyone ever seen this? I'm very puzzled as to what it might be. We'll do QC on the immortalised libraries tomorrow to see whether this has happen pre-capture or during capture, but thought I'd try here as well 🤔
*Thread Reply:* Hi Meriam. I think I'm seeing the same in a hyb-capture run I recently performed. I haven't figured out what this may be but looking at the sequencing data, the vast majority is off-target. Please let me know if you find out what's causing this 😬
*Thread Reply:* Would you mind sharing your BioAnalyzer run profile? It might still be “bubbles” but that will be very clearly visible on the plots
*Thread Reply:* That's what they look like!
*Thread Reply:* You are absolutely right. Doesn’t look like bubbles at all. How long are your adapters and are your fragments ligated to adapters prior to capture? Asking, just because a fragment with adapters of the expected size of 100bp is rather short
*Thread Reply:* I'm not sure exactly, but in previous batches we have fragments of about 160-170 bp in total on the bioanalyzer for an average size of about 60 bp of actual aDNA sequence. But these are 260, so another additional 100 bp...
I have a question for the community (this influences which pipelines we support for AMDirT - the AncientMetagenomeDir toolkit!):
Feel free to add other options 🙂
Hello @channel for those of you who are interested this is a reminder that tomorrow at 5pm CET we are having our first SPAAMtisch meeting! Chosen topic: what should SPAAMtisch be for you? If you have somehow missed it, you are still on time to join the #spaamtisch channel and feel free to be at the first meeting. Just to refresh the concept #spaamtisch will hold regular meetings for people who want to discuss some topics of interest chosen by the community and in an informal way! No knowledge or preparation required! All stress free 😉 The link to join below! https://gather.town/app/PlXjb0deog0B4JCq/spaam-community

hey @channel -- dont forget : SPAAMtish happening now!
#spaamtisch 👈:skintone3:more info
Hi @channel voting is open for the second #spaamtisch meeting date! Provisional date is the 12th of Jan 2023 at 5pm CET. Join the channel for more info and express your preference on the date! Cheers!
@channel last day to vote the date for the second #spaamtisch 👆
Does anyone know of other (student-like) initiatives like a bit like SPAAM but for other areas of biomolecular archaeology?
What about the sedaDNA people? Maybe @Eric Capo would know?
In the sedaDNA, we are organising student seminars once a month in the EU and North America, to cover most time zones. We sometimes have a theme, like shotgun sequencing or bioinformatics, and students share their experiences and can ask peers for help if they are stuck. Students and early postdocs from the SPAAM are welcome to join! there is a little more info on our website https://sedadna.github.io/category/events.html
*Thread Reply:* Does sedaDNA have any financial support or anything?
*Thread Reply:* No we don't have any financial support.
*Thread Reply:* Im going to message you 😉
@channel the second #spaamtisch meeting will be on the 12th of Jan 2023 at 5pm. Please go to the #spaamtisch channel to decide the topic to discuss if you are interested in participating and for more info about it🙂 Cheers!
Hello @channel hope you had a nice holiday time 🙂 this is a reminder for tomorrow #spaamtisch 12th of Jan from 5 -6 pm CET . We will chat about source tracking methods and if we have some times phylogenetic trees! Cheers!
@channel It's time to start preparing for the next SPAAM Summer School: Introduction to Metagenomics
Before I ask people directly, I would like to ask if anyone would be interested in volunteering to be an instructor for any of the topics (https://spaam-community.github.io/wss-summer-school/#/2022/README?id=schedule).
Note we will likely mostly re-use previous years material (unless someone wants to write their own/update), so your tasks would mostly be just presenting basic concepts and answer questions.
You will get official certification/proof of teaching that you can then use for CVs and other career portfolios.
We are looking currently for more experienced people (postdocs and upwards, to start). But may open up the call in the future (or if you're an advanced PhD and super enthusiastic you're welcome to volunteer already 😉 ).
If you're interested or have any questions, please contact myself or @Christina Warinner !
*Thread Reply:* do you already have a date in mind?
*Thread Reply:* Not yet, but around the same time - July/August (probably late/early)
*Thread Reply:* Is it online, or in person? i.e., do instructors have to be in Leipzig?
*Thread Reply:* Then I'd be happy to give a lecture, if you'd like! I could teach a number of the classes from last year - we can chat.
*Thread Reply:* Thanks! Yes, let's talk in real time
Hello @channel if you are interested please let me know via the poll on the #spaamtisch if the 16th of Feb at 5-6pm CET is a good date/time for you for our third SPAAMtisch meeting. Go to the channel for more info! Thanks!
*Thread Reply:* IMPORTANT: fill out the rota of topics if you want to propose something to discuss in coming meetings 🙂 (doc pinned in channel)
*Thread Reply:* Hey Maria, are these sessions recorded? I'd be very keen to watch them if there are talks/workshops etc. I'm in an awkward time zone 12 hours ahead of you 😬
*Thread Reply:* Hi @Meriam van Os nope atm we aren't recording them. However, we do take notes so you can read everything that has been discussed so far in the document pinned in the #spaamtisch channel.
Thank you to @James Fellows Yates and @Åshild (Ash) for our first SPAAM BLOG POST!!! If someone refers to a cluster, what on earth do they mean? Turns out it could mean all kinds of things! 🤯 But fear not, James and Ash break it all down for you here: https://spaam-community.github.io/blog/2023/01/23/blog-cluster/ Coming up next: a fantastic crowd-sourced explainer from @Kevin Daly on how deep to sequence to capture 🦠🦠🦠 Have something you’ve just figured out and want to share? Have a question you want to crowd-source the community’s wisdom about? Just want to be ✨published✨? Hit up @Ele and me with your ideas!
*Thread Reply:* Don't forget to tweet about it!
ISBA10 Abstract submission is open until March 15th :mask_parrot: https://www.isbarch.org/events/news/2023/01/25/events-isba10-abstracts-open.html https://isba10.ut.ee
Hola @channel, If if you are interested in sharing information on funding opportunities 💸 for postdocs or junior PIs and discussing suggestions for a successful outcome, please join to <#C04MPAVC83F|early-career-funding-opportunities> :spaam:!!!
Hello @channel the time of the third #spaamtisch meeting is approaching (16th of Feb 2023 5-6pm CET) so please express your preference on the discussion topic and please N.B. you can add an option in the poll (e.g. if you would like something more introductory to be discussed on some particular topic). Also, don’t forget we’ll have a guest speaker in March :partyparrot:
*Thread Reply:* Hybrid event is not an option you consider?
*Thread Reply:* Hi Jasmin, we were more thinking of a discussion based format for the in-person meeting (maybe some “round tables”). We might have a camera set up but don’t know for sure yet.
Hello @channel our next two #spaamtisch meetings will be in different formats. I have invited two authors presenting their published or nearly published recent research work. For our first #spaamtisch of this format we’ll have @Camila Duitama presenting on the 21st of March at 5-6pm CET. Camila will present her work on:
decOM: Similarity-based microbial source tracking of ancient oral samples using k-mer-based methods https://www.biorxiv.org/content/10.1101/2023.01.26.525439v1.full
Everybody can participate to this meeting. However, as per community vote earlier this month, PIs will be allowed to ask one question in order to give the opportunity to young researchers and students to be more involved. So read up and prepare your questions! 🙂

@channel Please don't forget that the ISBA10 abstract submission closes next week (March 15th) ! We look forward to your submissions 😀 https://isba10.ut.ee/abstract-submission
*Thread Reply:* > "Abstract submitted successfully" :toad_scream:
*Thread Reply:* just to clarify -- can we still submit on the 15th? if so, when is the cutoff time?
*Thread Reply:* At 23.59 CET on the 15th March.
*Thread Reply:* and should we indicate anywhere that it could be ideally a talk but also considered for a poster?
*Thread Reply:* see post in the main chat 🙂
@channel Also, you could send an abstract to the SMBE too! Especially to this symposium, "Host-pathogen co-evolutionary dynamics through the lens of paleogenomics." https://www.smbe2023.org/abstractsubmission. The deadline for abstract submission will be on the 15th of March too!! 🦠
Is it the year of red colour schemes and abstract deadlines on the 15th of march? 🤔
*Thread Reply:* Wait isn't that the ides of march too 😶🌫️
*Thread Reply:* https://www.pantone.com/color-of-the-year/2023 close enough? 👀
Just some things to clear up because they aren't stated clearly on the website and there was some miscommunication across disciplines: • Abstract deadline is 23.59 CET on the 15th March • All talks will also be considered for posters • There will be an extension announced later today This will all be updated on the website asap
@channel Its official: ISBA10 Abstract Submission Deadline extended to 23:59 CET on 31st March 2023! Info has been updated on the website: https://isba10.ut.ee/abstract-submission
*Thread Reply:* quick q - what's the connection between SPAAM and ISBA, is there a meeting coinciding with the conference in September? thanks!
*Thread Reply:* Hi, Yes tentatively a meeting a day before ISBA10 is planned in Tartu. It is currently still in planning. But you can check the general chat for a poll regarding preference we sent out a couple of weeks ago.
Hello @channel don’t forget our meeting today at 5-6:30pm CET with @Camila Duitama presenting her PhD work on ancient metagenomics decontamination.
Find below the link to the Teams meeting. You do not need a Teams account or app installed on your device if you just choose the option “Join from web” on Google Chrome as your web browser. See you later!!!
Microsoft Teams meeting
Join on your computer, mobile app or room device
*Thread Reply:* I'm wondering if there could be a video recording, because it will be already midnight in China at the time of the meeting.🫣
*Thread Reply:* Hi @Wenqin Yu btw 😉 👋
*Thread Reply:* Hi @Wenqin Yu yes there will be a video recording that I will upload sometime soon on the youtube channel of spaam 🙂
*Thread Reply:* That's pretty good, thanks!
@channel we are online now 🙂 👆 in few minutes Camila will start presenting
Dear @channel, today is the last day for ISBA10 abstract submissions. This time it's the real deal! 👀 So if you are still planning to submit something, please do it by 23:50 CET on March 31st 2023. https://isba10.ut.ee/
Aaaaaaand apparently I forgot to press enter on this announcement 🤦 , here it is...
🚨 Applications for the SPAAM Community's Introduction to Ancient Metagenomics Summer School 2023 are now open!🚨
*Date and schedule*: Online from Monday 31st July to Friday 4th of August from 09:00 to 17:00 (CEST, Leipzig time zone).
*Instructors*: Dr. Aida Andrades Valtueña, Dr. Alexander Herbig, Alina Hiß, Dr. Arthur Kocher, Clemens Schmid, Dr. Kevin Nota, Dr. Meriam Guellil, Dr. Nikolay Oskolkov, Robin Warner, Dr. Thiseas C. Lamnidis (more TBC)
*Course Overview*: Ancient metagenomics applies cutting-edge metagenomic methods to the degraded DNA content of archaeological and paleontological specimens. The rapidly growing field is currently uncovering a wealth of novel information for both human and natural history, from identifying the causes of devastating pandemics such as the Black Death, to revealing how past ecosystems changed in response to long-term climatic and anthropogenic change, to reconstructing the microbiomes of extinct human relatives. However, as the field grows, the techniques, methods, and workflows used to analyse such data are rapidly changing and improving.
In this hands-on summer school (block praktikum) we will go through the main steps of ancient metagenomic bioinformatic workflows, familiarising students with the command line, demonstrating how to process next-generation-sequencing (NGS) data, and showing how to perform de novo metagenomic assembly. Focusing on host-associated ancient metagenomics, the course consists of a combination of lectures and hands-on exercises, allowing participants to become familiar with the types of questions and data researchers work with. Round table discussions with experts at each stage of the workflow will be held to allow participants to get advice on their own projects and research.
By the end of the course, participants will have an understanding of how to effectively carry out the major bioinformatic components of an ancient metagenomic project in an open and transparent manner. Attendees will be eligible for ECTS points (awarded by the JSMC Graduate School, Friedrich-Schiller University, Germany) or a certificate of completion (Max Planck - Harvard Research Center, MHAAM).
**Eligibility: The course is aimed at masters students and early-stage PhD students, to a maximum of 25 participants. Course instruction will take place online.
*Applications*: Applications are open from April 3 to May 26 2023. To apply, please visit our website: https://spaam-community.github.io/wss-summer-school/
Wow, looks awesome! Though time zones are gonna be a problem (starts at 3 AM for me 🥴). Are there similar opportunities available in American time zones?
*Thread Reply:* Currently not, sorry. At least not from us. I would love to be able to run it in different time zones, and I've constructed the material so other SPAAMers/people can run the same course elsewhere.
I'm happy to act as a consultant if you can get enough teachers in your timezone
Note all the material and recordings go online under a creative commons license so you can also follow outside of the summer school itself
*Thread Reply:* Thanks, I'll probably follow along with the recordings, then. Hopefully there will be momentum to teach it live in other time zones soon!
*Thread Reply:* The more people ask loudly the more likely it will happen! ;)
On behalf of the organizing committee, I am happy to announce that the abstract submission for the upcoming ISBA10 Meeting is closed. We have received nearly 350 abstracts. Thank you all for sharing your research and planning to come to Tartu. We have sent out a confirmation letter to all whose abstracts we have. If you did not receive a letter, but you did submit an abstract for the ISBA10 Meeting, please contact the organizing committee: isba2023@ut.ee. During April the assigned scientific committees will evaluate your abstracts and at the beginning of May we will send out the results. We aim to get the final program ready in June.
✨ New SPAAM blog post up!! 🧬 @Kevin Daly wants to detect pathogens in ancient livestock — how many reads should he shoot for? 🐄 He asked for wisdom from the wonderful SPAAM community and wrote up the crowd-sourced advice here: https://spaam-community.github.io/blog/2023/04/07/blog-numreads/ 🦠 Congratulations Kevin and thank you for your contributions!!
@channel If you submitted an abstract to ISBA10 but got error messages and then never received a confirmation email, please send an email to the ISBA10 email and cc the head of the committee you submitted the abstract to, There have been a few cases apparently. You can also DM me.
*Thread Reply:* Is registration for ISBA10 open yet? There doesn't seem to be any links on the website.
*Thread Reply:* It will be open soon, there was some delay because of the abstract extensions and website developer. I will announce it here once it is live but it should be soon
Dear @channel save the date! As anticipated our next #spaamtisch meeting will be in seminar format and it will be done on the 18th of April 2023 at 5-6 pm CET via Teams. Our next speaker will be @Andrea quagliariello who will present his recent work on ancient oral microbiome. More info below.
https://www.nature.com/articles/s41467-022-34416-0
Dear @channel don’t forget tomorrow the 18th at 5-6pm CET our second seminar for SPAAMtisch where @Andrea quagliariello will present their recent work!
Find below the link to the Teams meeting.
You do not need a Teams account or app installed on your device if you just choose the option “Join from web” on Google Chrome as your web browser. See you tomorrow!!!
Microsoft Teams meeting
Join on your computer, mobile app or room device
*Thread Reply:* Thank you so much @Maria Lopopolo for arranging the SPAAMtisch seminars! Is there any chance that they start a bit earlier during the day? I typically have family commitments at 5-6 pm and miss the talks every time 😞 I understand that times won't be optimal for everyone in any case, but perhaps alternating would be an option?
*Thread Reply:* Hi Nikolay, I am sorry to hear that 😞 . I can try to do a poll with some options to see how to better accommodate the majority. But the the main reasons we do that time are 1. Because many people who aren't at CET, the earlier we do it the less would be involved and 2. Even the people at CET often prefer to attend/be available for seminars after "official" working hours. However, for those that you have missed, I am recording the talks and they will be uploaded on the YouTube channel in the next few days. Nevertheless, I'll try to see what I can do for next month ;)
*Thread Reply:* Cool, I did not know about the recordings, would be glad to watch some 🙂
Hello everyone!
As #spaamtisch looks like it's quite popular (>30 people joining each session), we would like to find someone to support @Maria Lopopolo in the organisation (mostly inviting speakers, planning session topics, and making sure a video call link is available).
Is there anyone interested in getting involved in the community in such a way? (Maria can correct me if I'm wrong), but I think the tasks only takes about 1h a month max. We just want to make sure there is someone to ensure each even goes ahead (e.g. if Maria is on holiday etc.). If you're interested please contact Maria or myself 🙂
*Thread Reply:* Yes and I take the opportunity to thank @irinavelsko and @Miriam Bravo who helped me moderating the two last sessions of the seminar format #spaamtisch. I'd like also to remind that these meetings will also keep alternating with the "regulars' table" more informal format where we discuss a community chosen topics :)
Hello Everyone!
We are thrilled to announce that registration for the first in-person SPAAM meeting since the pandemic is beginning, SPAAM5! This year we are holding a one-day only in-person meeting on September 12th, 2023 in Tartu, Estonia. We are taking advantage of this year’s ISBA meeting in the same location to meet in person the day before it starts. As attendance is limited, we will give priority to early career researchers. This meeting will feature networking, flash talks, and more! But if you can’t make it, don’t worry, another online-only meeting will be held in November to make sure as many people as possible can participate in SPAAM5. Additionally, anyone who cannot present in person during the Tartu meetings this September will be prioritized for online-only presentations in November.
To register for the in-person event, please fill out this form by June 16: https://forms.gle/nJUMGzTmH6Knuy7R9.
For selected participants, a 15 Euro registration fee will be collected prior to the start of the meeting and will cover lunch, coffee and refreshments. Registration waivers are available upon request: please contact spaam.events@gmail.com.
Looking forward to finally meeting in person,
The SPAAM5 Organizers
*Thread Reply:* Hey @Zoé Pochon when will we know if we have been accepted/there is space for us?
*Thread Reply:* We've now got ISBA acceptance emails and will need to start booking stuff already
*Thread Reply:* (so need to know whether can factor in a day early or not)
*Thread Reply:* Good question, let me check with the others. For the moment there is only like 12 people registered so I hope we won't have to do a selection
*Thread Reply:* Might be worth sending Reminders everywhere now acceptance emails are out and they have a better idea if thy are going or not :)
*Thread Reply:* Yes! I suggested it to the others, we will do that as soon as possible 😉. Thanks James 😊
*Thread Reply:* You could also maybe ask ISNA people to mention it in one of their emails
*Thread Reply:* You could also maybe ask ISNA people to mention it in one of their emails
@channel you're getting an extra reminder ;()
Registrations for the SPAAM ancient metagenomics summer school is closing NEXT FRIDAY! Please share the word and your colleagues/students :)
https://spaam-community.slack.com/archives/CPYE64ZC6/p1680521393090799
@channel Reminder to register now for SPAAM5 - there are still free spots for the SPAAM meeting in Tartu, Sep 12 (one day before ISBA starts). For questions contact spaam.events@gmail.com or the organisers: @Alina Hiss @Megan Michel @Sierra Blunt @Olivia @Zoé Pochon @Alice Lee @Meriam Guellil https://spaam-community.slack.com/archives/CPYE64ZC6/p1682519247837749
Very last reminder for the summer school applications! The Submission form closes tonight!
https://spaam-community.slack.com/archives/CPYE64ZC6/p1680521393090799
*Thread Reply:* @James Fellows Yates I do not recall (and can’t find) if you sent any email via SPAAM mailing list that I could share with my colleagues in case somebody wants to register last minute?
*Thread Reply:* Nevermind, I can just copy the text from the announcement above and send as a separate email to my colleagues 🙂
*Thread Reply:* I sent multiple to the SpAAm mailimh list
*Thread Reply:* Are you not on it
*Thread Reply:* Oh, I think I found it, sorry! 🙂 I checked thus morning on my scilifelab address but the one included in SAAM mailing list is probably my Lund University email 🙂 Sorry for my confusion!
*Thread Reply:* You can always sign up with the other is your wish
*Thread Reply:* And we can cancel the lund
*Thread Reply:* No worries, I use both of them. I was just for some reason sure this morning that SPAAM is using my scilifelab email 🙂
*Thread Reply:* Hi there! Wondering if there was any news about the summer school for those of us who applied? 😬 Colleagues and I haven’t heard back and wondering if no news is bad news haha. Thank you!
Hello @channel today there is our #spaamtisch meeting at 5-6 pm CET! We will discuss together about:
- Functional analyses on pathogens or microbial communities
- If we have time: Taxonomic classifiers! We will gather in gather.town (see link below) See you soon!! ☺️ ☺️
Hi! I will upload the notes from the meeting! If you are not in the channel of the #spaamtisch join it so you can check the notes (pinned in the channel) 😊
Just saw this in my inbox and they have a specific section in the section for Ancient DNA... I guess they are looking for input, if anyone is interested
```Dear ENA Users,
The ENA has designed a Data Use Survey to gain better understanding of our users who access and download data from the ENA (and/or other repositories) and their experience using our services. We hope to make use of any feedback received from this survey to understand and improve user experience.
We would greatly appreciate it if you could take the time to fill out and submit this survey linked below:
ENA Data Use Survey
It should take approximately 9 minutes to complete.
Kind Regards, The ENA Team```
FYI, github changed some permissions things and I'm having to fix some stuff, so for everyone in the GitHub organisation, sorry if you get a couple of emails in the next hour!
Dear @channel as promised we have published the two seminars (@Camila Duitama here and @Andrea quagliariello here )from our previous SPAAMTisch meetings on our community youtube channel! Do catch up with these amazing seminars if you were not able to follow the live meetings!
Hi @channel if you want to attend the next #spaamtisch seminar given by our very own @Maxime Borry and @James Fellows Yates on AMDirT, a poll is waiting. Go on the #spaamtisch channel and choose the dates that work better for you!Cheers!
The SPAAM twitter account has more than 500 followers now. :partyparrot: https://twitter.com/spaam_community

For those asking about the summer school: we had 100 applications that Tina and I need to go through! The decision emails will be sent out within a couple of weeks!
Hi all, has anyone heard about ISBA bursaries? Trying to think through travel plans and I’m fairly sure I didn’t receive anything about it, but the website says that was in April/May 😕
*Thread Reply:* I can ask back channels for you for an informal answer
*Thread Reply:* Also I'm sure members of the ISBA conf committee are also watching 😉
*Thread Reply:* we are 😅 but I am sadly also not knowledgeable about this (I have asked a bunch of times the last couple of months but never gotten an actual reply) but I have reached out yesterday again. If I hear back I will let you guys know.
@channel Reminder to register for SPAAM5 until this Friday 16th of June - the SPAAM meeting will take place in Tartu, Sep 12 (one day before ISBA starts). For questions contact spaam.events@gmail.com or the organisers: @Alina Hiss @Megan Michel @Sierra Blunt @Olivia @Zoé Pochon @Alice Lee @Meriam Guellil https://spaam-community.slack.com/archives/CPYE64ZC6/p1682519247837749
Today is your last chance to register to the SPAAM event, which will take place in Tartu, one day before ISBA, on September 12th! Looking forward to seeing you there 😊!
Hello @channel according to vote and availability of the speakers, the selected date for the next #spaamtisch meeting will be on the 26th of June at 4-5pm CET. One hour before our usual time. The meeting will be a seminar format where @Maxime Borry and @James Fellows Yates will present our community paper on AMDirT! See you soon! Link to Zoom will be sent closer to the date. Cheers!
Dear @channel don’t forget on the 26th of June at 4-5pm CET there is going to be our next #spaamtisch meeting! It is going to be in seminar format , where @Maxime Borry and @James Fellows Yates will present the community paper on AMDirT! See you soon!☺️ Find below the link to the Zoom meeting.
Biancamaria Bonucci is inviting you to a scheduled Zoom meeting.
Topic: SPAAMtisch meeting Time: Jun 26, 2023 04:00 PM Amsterdam, Berlin, Rome, Stockholm, Vienna
Join Zoom Meeting https://ut-ee.zoom.us/j/98167778236?pwd=SW9wMzVUY1I2ZXRFclN5bmJRSm9tZz09
Meeting ID: 981 6777 8236 Passcode: spaam
Hey everyone, For those of you planning to go to ISBA in September, don’t forget that the early bird registrations ends up next week (30th of June). On a similar note, I’d be happy to take any recommendations for a good restaurant as an alternative dinner party to the gala dinner ;)
*Thread Reply:* I'm planning to exploit some friends there to go to somewhere instead too 😬
*Thread Reply:* I am free to join you, too 😁
*Thread Reply:* I just registered with Nils (from HR) paying the conference fee. He is asking if you @James Fellows Yates want to register next week Wednesday as well or did you pay yourself already?
*Thread Reply:* I've paid already myself
*Thread Reply:* I was told they only do reimbursements not direct payments on your behalf
*Thread Reply:* From Nils' question I assume that @Maxime Borry still hasn't filed the Dienstreiseantrag? 😬 PM me if you need help from someone physically at HKI
*Thread Reply:* They pay on our behalf ONLY conference fees, no travel expenses/hotels beforehand.
*Thread Reply:* Wewll it would be nice if they documented this but oh well
*Thread Reply:* @Jasmin Frangenberg no, i haven't done it yet
Dear @channel This is to remind you that today at 4-5pm CET there is going to be our next #spaamtisch meeting! It is going to be in seminar format , where @Maxime Borry and @James Fellows Yates will present the community paper on AMDirT!
See you later!!!☺️ Find below the link to the Zoom meeting.
Biancamaria Bonucci is inviting you to a scheduled Zoom meeting.
Topic: SPAAMtisch meeting Time: Jun 26, 2023 04:00 PM Amsterdam, Berlin, Rome, Stockholm, Vienna
Join Zoom Meeting https://ut-ee.zoom.us/j/98167778236?pwd=SW9wMzVUY1I2ZXRFclN5bmJRSm9tZz09
Meeting ID: 981 6777 8236 Passcode: spaam
*Thread Reply:* hello! Is there a recording of a seminar for the pacific time zone folks? cheers : )
*Thread Reply:* We did record it so will go up soon!
Just to let you know the SPAAM website will be done for a day or so! So please don't worry if you can't access it currently 🙂
It's back up for now but may go down again this evening for a bit
Hello all, Thought someone can help me with an ISBA related question. If I'm registered but can't make it to the conference due to visa issues, do I get a refund? Couldn't find a contact address on their website.
*Thread Reply:* Please contact the organising committee at the email address provided on the website: Organising committee: isba2023@ut.ee
*Thread Reply:* Not sure about the answer though since I believe this would be the first (although maybe not the last) case of this sort. This will depend on the Estonian side.
*Thread Reply:* But hopefully it can be sorted. I have given them a heads up.
*Thread Reply:* I was just told a refund will be available but please get in contact with the email admin
*Thread Reply:* Yes they just told me the same thing. Thanks again
Reminder if you're joining the jumping of ship 🐦
You can help ancient DNA researchers reconstruct their networks (and find new interesting follows) by adding yourself to the following list: https://genomic.social/@jfy133/110648759653397397
🚨Hello @channel🚨 📣Check out the FIRST SPAAM :spaam: NEWSLETTER!! 📣 :headbangingparrot: 🚨We are thrilled to bring you an exciting update on the community, upcoming events, current and new initiatives, recent publications from the past three months, and available job positions. 🚨 We will post the next one in three months! :spaam::catjam:
Loving the :doggo: ❤️ in the steering committee 😉
Dear @channel summer is here! July #spaamtisch session will be our cosy “regular’s table” format :spaamtisch: Please reply to the two polls posted on the #spaamtisch channel for the date and the topics you would like to discuss.
Also please don’t forget to populate our “rota of topic” document pinned in the #spaamtisch channel so that you get the chance to discuss your topic of interest too!
Spaamtisch will likely take a break in August due to summer holidays. However, we will most likely send out a survey for you to fill out so that we can get your opinions into consideration to make spaamtisch more useful for everyone for the next academic year :spaamtisch: Cheerio!
*Thread Reply:* cheerios
@channel Hi Everybody! A few of us have been discussing having a writing group where we can block off a couple of hours and zoom together while we each focus on writing whatever manuscript, abstract, proposal, etc. we are working on. This would be a group writing block where we can each just create some accountability and support for dedicating time to write. The name of our writing group has been granted by none other than James Fellows Yates - SPAAMFIC 😎. I’ll be sending out a poll to see who is interested in joining SPAAMfic and add you to our channel!
*Thread Reply:* Sooooooo......
Hi @channel, SPAAM has a new URL!! you can find it easily at https://www.spaam-community.org ! and do not worry about old links.. the github still works and will direct you automatically to the new url. (thanks to @James Fellows Yates for setting this up!)
Hello @channel! As mentioned in Newsletter #1:catjam:, we're excited to announce that we have crafted a Constitution :spaam: (https://www.spaam-community.org/constitution/). We wholeheartedly encourage you to share your thoughts, suggest changes, or provide any feedback you may have 💬. Please feel free to reach out to any member of the steering committee (@Gunnar Neumann, @Shreya, @Maria Lopopolo, @aidanva, @Kadir Toykan Özdoğan, @Ian Light and me). We greatly appreciate your involvement! 🙂
@channel in case anyone is interested to join, we are offering a course this fall in ancient environmental genomics: https://twitter.com/miwipe81/status/1671094458039820289?s=46&t=gwKWZJ5qldtF98LPMY9EQ|https://twitter.com/miwipe81/status/1671094458039820289?s=46&t=gwKWZJ5qldtF98LPMY9EQ

Hey all!
If you saw on the newsletter from the steering committee, we have a new SPAAM project: the Little Book of Smiley Plots
The project aims to aggregate all types of damage plots, and describe what causes them, to make it for you to interpret different plots as they come up in your own data. To make it more 'fun' we are making caricatures of each plot (see existing exmples in the book uidner 'no-UDG double-stranded' and 'insufficient reads'.
https://www.spaam-community.org/little-book-of-smiley-plots/
So we need two sets of contributions:
- Weird damage plots
- Artistic representations/cariactures of the plots We currently have three different smiley plots on the website without a caricature 😞 so if you're feeling creative, please join <#C05F0URFJ7M|little-book-smiley-plots> and shout if you want to join in 😄
Note that we want to leave as much creative freedom as possible, (only a couple of specifications), and quality is not important!
Cheers, James and @aidanva
*Thread Reply:* Here is a weird contribution, that took me a while to find out ! Answer tomorrow evening 😉
*Thread Reply:* Yes please 😄
*Thread Reply:* Ligation biases to barcoded adapters (double-stranded library).
*Thread Reply:* Ooooh! Is that a published sample? And can you write a short blurb
*Thread Reply:* Yes, that's from https://doi.org/10.1093/molbev/msac263 but I don't think the plot itself is included in the publication. Similar (possibly more obvious) plots are included in the supp mat here (https://doi.org/10.1093/molbev/msaa135, Fig. S11) and the explanation for the pattern is in the legend and the supp methods text. "During this investigation, we observed unusual damage patterns in several of our samples – rather than the expected incremental rise in deamination towards read ends, several samples showed a drop in the frequency of C-to-T substitutions from the penultimate base to the terminal base (e.g. comparison of damage patterns in Ua9 MAGs compared to Ua14 in Supplementary Fig. S11). It has been suggested previously that barcodes with a terminal G or C have reduced ligation efficiency to damaged cytosines at 5’ ends of reads (Rohland et al. 2015). We therefore examined damage profiles for 14 most abundant bacterial taxa in the post-filtering dataset (Supplementary Table S6), using mapDamage as part of the EAGER pipeline v1.92.37 (Peltzer et al. 2016). We only considered cases with more than 10000 reads mapping to a taxon reference genome in a given sample in order to have enough coverage to discern a clear pattern above the background noise. We found that samples containing barcodes with a terminal G or C were more likely to show the unusual pattern (χ2 = 31.11, df = 1, p-value < 0.0001) (Supplementary Table S13)."
*Thread Reply:* I can try and get the data from Ena and try to recreate
*Thread Reply:* Unless you have the mapDamage files still
*Thread Reply:* Was trying to dig them up and cannot find them. Maybe @Jaelle Brealey @Jaelle Brealey (not sure if she's active here and under which of the two handles) could be of help
*Thread Reply:* Will try to reach her elsewhere
*Thread Reply:* I should be able to pull up the mapDamage files somewhere, I'll dig through my old data folders.
*Thread Reply:* For the one I posted above, it was soft clipping because of using bowtie2 in end-to-end alignment mode 😉
*Thread Reply:* @Maxime Borry could you supply me a bam file so I could make a mapDamage/DamageProfiler plot? (alternatively the pyDamage raw files so I can partly recreate your plot)
Hi all, The SPAAM community has a Mastodon account now! :partyparrot: You can follow it here: https://genomic.social/@spaam_community
FYI, as we are currently in a 'free trial' of slack, I'm making an export of all our slack data (that we can), that way we can review old messages if you ever need!
I will keep a back up as will the steering commitee 🙂
Hi everyone!!! As previously mentioned, we prepared a survey for you to fill out, so that we can collect feedbacks and opinions on #spaamtisch! We will take answers till the 31st of August. We can’t wait to see your feedbacks, in order to make the event a better experience for everyone!!
And, if you don't know what #spaamtisch is...go check it out in the slack channel!:catjam: Cheers :spaamtisch::spaamtisch:
Hiya SPAAM Community!
We are super excited that, as the Steering Committee and the SPAAM5 organizing committee, we have put together a SPAAM 5.0-themed T-shirt!
There are two styles, one with the design only on the back of the shirt, and one style with the same design on the back, but also a small SPAAM logo on the front. The shirts are available in a variety of colors.
Here are the links to the ordering website: Dark styles, back of shirt design only Light styles, back of shirt design only Dark styles, front+back Light styles, front+back
We hope that you can get a shirt if you are interested, even if you don't come to SPAAM 5.0!
Please note: if you wish to purchase a shirt, you are responsible for shipping. If you are attending SPAAM5 in Tartu, and wish to have the shirt before that, you should order ASAP! If for whatever reason ordering a shirt if you reside outside Europe is not possible and you are coming to SPAAM5.0, please contact us and we may be able to help!
Best, The Steering Committee of SPAAM (Aida, Betsy, Gunnar, Ian, Kadir, Maria, Miriam, Shreya)
If anyone has trouble ordering the Shirt to their country but is coming to the SPAAM5 meeting and/or ISBA, let me know and we can try to arrange sth.. i can probably fit some extra shirts in my luggage.
What, this shirt ;)?
(Don't let the model put you off, the shirt is pretty good! Note they run on the slightly larger/longer side, at least medium)
*Thread Reply:* Thanks for sharing that info 🤏
*Thread Reply:* Is it still possible to order? I tried and amazon said that it has been already sold 😞
*Thread Reply:* Strange... should be on demand AFAIK?
*Thread Reply:* @Kadir Toykan Özdoğan maybe will need to investigate
*Thread Reply:* This is what I get. Hmm, perhaps it does not explicitly say “sold” but something did not work
*Thread Reply:* Isn't the adress the problem there?
*Thread Reply:* Oh, it might be the location.
*Thread Reply:* I put my Swedish address 🤔
*Thread Reply:* It is possible that they do not send it to Sweden, although would be strange.
*Thread Reply:* Maybe they do not like ä, å and ö Swedish letters 🙂 I will try again
*Thread Reply:* I can’t get one to ship to the UK either 🥲
*Thread Reply:* @Gunnar Neumann maybe you should come up with a procedure/instructions how others can order via you?
*Thread Reply:* This is easier to solve @Kelly Blevins. I can make them available for the Amazon UK, should just take a couple of hours.
*Thread Reply:* Is that beard for sale also? 👀
*Thread Reply:* assuming it wont be a huge mass of people having issues, people can just contact me directly and we discuss deatails. I would put the order at the end of the month, September 30th
*Thread Reply:* UK links: only back print https://www.amazon.co.uk/dp/B0CGJ4ZBLM https://www.amazon.co.uk/dp/B0CGJ65RX1
back&front https://www.amazon.co.uk/dp/B0CGJ6NFK4 https://www.amazon.co.uk/dp/B0CGJDXN27
*Thread Reply:* It doesn't work to order it to Sweden for me either
*Thread Reply:* Maybe for Sweden people you can send the money to Gunnar to order for you then?
*Thread Reply:* Would that be okay @Gunnar Neumann? I can transfer you the money (Revolut, bank transfer... )
*Thread Reply:* Of course @Zoé Pochon just let me know what size, colour , print (back only or back/front) Maybe you can just pay in cash in Tartu
Hi all!
For those *not* going to the ISBA gala dinner, if you're interested in maybe joining a group of us to go to a restaurant somewhere else, please react to this message with a :knifeforkplate: .
If it's not too many people we will maybe try and reserve table somewhere!
Good Morning, @channel, last call for those who had issues ordering the SPAAM5 Tshirt. I will place an order tomorrow morning and could bring the shirt to Tartu in two weeks…
*Thread Reply:* @Gunnar Neumann The Amazon page says they are all out of stock. Do you have suggestions?
*Thread Reply:* I’d like to order a men’s large navy shirt
*Thread Reply:* @Tina Warinner looks fine here when i just checked.. i will order one for you then, too. Will you be in Leipzig before? otherwise i bring it to Tartu.
*Thread Reply:* @Gunnar Neumann I have the same question. If there are some left, I’d like to order a men’s medium black shirt.
*Thread Reply:* Oh, wait I fixed it. My shipping address was set to the US and it didn’t work, but now that I changed it to Germany it works
*Thread Reply:* I’ll go ahead and order it online to deliver to the institute. Thanks @Gunnar Neumann!
*Thread Reply:* ok.. i take you off the list then.
*Thread Reply:* sure, @Sarah Johnson i will order one for you. they will be printed on demand/order.. so there are as many “left” as we want. 😉
*Thread Reply:* i assume you want print on back and front?
*Thread Reply:* @Gunnar Neumann I see! That’s great thank you so much. Yes I would like it printed on both back and front. Let me know how I can send you money. I’ll be in Tartu beginning the 12th.
*Thread Reply:* you can pay me in cash on site.. otherwise we figure something out when there..
*Thread Reply:* Thanks a lot @Gunnar Neumann for arranging it!
👀 anyone gonna be in Tartu next Monday afternoon and want to do this adventure park with me? http://www.tartuseikluspark.ee/ Nothing puts pre-conference nerves in perspective like dangling from a rope ladder 12m off the ground 😃

*Thread Reply:* It’s really close to the conference venue, so depending on when SPAAM wraps up we could potentially go on Tuesday if that works better for people
*Thread Reply:* I'm in!!
*Thread Reply:* I will arrive in Tartu on Friday, but I can't join any "sports" atm. Happy to join you for a coffee or something else on Monday afternoon/evening.
*Thread Reply:* 🥺 I'd love to, but will prbly not arrive before 7pm. But let me know your experience! Because I will have plenty of time after the conferences (6 days vacation).
*Thread Reply:* @Tina Saupe I'll be arriving in Tartu around 17 (they booked me a later train than expected). A coffee sounds great to me since as it turns out I may be getting in too late for the adventure park. (Sorry @Kelly Blevins 🙁)
*Thread Reply:* No worries! My train gets in around 3. I will go adventure parking after that. But perhaps we could meet up for drinks/coffee later this evening?
*Thread Reply:* Coffee/drinks sounds fabulous!
*Thread Reply:* I’m https://maps.app.goo.gl/XXuXDTD3wBSXc5WR6?g_st=ic if anyone wants to join 🍻
HIVE-MIND ASSEMBLE!
WHAT ARE YOUR FAVOURITE OR GO TO INTRODUCTORY/GENERAL OVERVIEW REVIEWS OF ANCIENT METAGENOMICS AND RELATED ???
Post citations in the thread, if your favourite is already posted then give it a thumbs up
*Thread Reply:* SPAAM Blog of course!! 😉
*Thread Reply:* Well that's a given, but I meant more proper publications
*Thread Reply:* https://pubmed.ncbi.nlm.nih.gov/28460196/

*Thread Reply:* https://www.nature.com/articles/s41576-019-0119-1
*Thread Reply:* https://www.nature.com/articles/s43586-020-00011-0
*Thread Reply:* On a roll @Camila Duitama :catjam:
*Thread Reply:* I guess I have reading for the plane on the way to SPAAM-5
*Thread Reply:* Let us know your favourite after @Dawn lewis!
Hey all! I was asked to give the "Metagenomics" invited talk at ISBA, and yet I find myself wondering if I even know what Metagenomics is! Can I tap the hive-mind and ask for your opinions on a few questions?
I think a main challenge for me is that I think of Metagenomics as "studies that use samples with DNA from many organisms / taxonomic groups." But I think a more accurate statement is "Metagenomics is the study of communities of organisms via DNA."
More specifically: Do Metagenomics studies have to study contemporary organisms - i.e., does that have to be the goal? If you're studying dental calculus, you're presumably hoping to get a community of organisms that all lived on the tooth at the same time, and not the stuff that colonized it later.
*Thread Reply:* Two person consensus from myself and Irina: to answer your questions in order:
- "studies that use samples with DNA from many organisms / taxonomic groups." Not necessarily
- "Metagenomics is the study of communities of organisms via DNA": Yes
- "Do Metagenomics studies have to study contemporary organisms": No
- "Is a study that uses metagenomic samples (i.e. sediments), but focuses only on one species / taxa within those samples still a metagenomic study?": No Longer form:
The main issue here for you is that all 'ancient' studies are interestingly metagenomic. They always have a community of organisms ancient/modern. In most cases all ancient DNA studies will use metagenomic 'approaches' or consider concepts that are a strong focus of metagenomics (e.g., contamination from other organisms). A metagenomics study does not have to be studying purely contemporary organisms - you also have necrobiome studies, sediment studies over a time transect is still metagenomics, if you are aiming to look at the whole or large fraction of the community of organism.
Specifically for 4: these studies indeed apply a little bit of metagenomic approaches or techniques initially, however the end goal is ultimately indeed not 'metagenomics' given the strong focus on single taxa.
*Thread Reply:* In our opinion
*Thread Reply:* This is really great, thank you! It helps to clarify my thinking a lot. Essential, clarifying between metagenomic data/approaches and metagenomic STUDIES.
*Thread Reply:* Agree with James and Irina.
To add re. definition of metagenomics: I think both are correct — the first for metagenomics methods, the second for metagenomics analysis/interpretation. I would add ‘genome-wide DNA’ though (to separate from metabarcoding).
As a [descriptor] study is based on the approach used for analysis/interpretation, community-reconstruction sediment and microbiome studies can be called metagenomic. IMO.
*Thread Reply:* Also very excited for whatever Pete is typing 🙂
*Thread Reply:* >This is really great, thank you! It helps to clarify my thinking a lot. Essential, clarifying between metagenomic data/approaches and metagenomic STUDIES.
This is what I should have said back in that late tired evening ;)
*Thread Reply:* Ah, so you would say that metabarcoding studies are not metagenomic?
*Thread Reply:* I would argue not. They share the ‘meta’ aspect, but they are not ‘genomic’
*Thread Reply:* > Ah, so you would say that metabarcoding studies are not metagenomic?
*Thread Reply:* Absolutely not
*Thread Reply:* They are Metataxonomic
*Thread Reply:* Wait ignore the absolutely before I put my foot in it
*Thread Reply:* metataxonomic, I like that
*Thread Reply:* That's a true term!
*Thread Reply:* I promise not to screenshot any more of your replies, James!
*Thread Reply:* but surely metagenomics is metataxonomic, too?
*Thread Reply:* More recent papers describe it
*Thread Reply:* > but surely metagenomics is metataxonomic, too? > Yes, it's just not bidirectional
*Thread Reply:* > I promise not to screenshot any more of your replies, James! > I dunno, maybe we can make it a tradition ;)
*Thread Reply:* Ok, followup question: Are metagenomic studies primarily interested in taxonomic abundances and/or presence/absence?
*Thread Reply:* Not anymore. It's often a first component of a metagenomic study, but it's less and less the sole focus
*Thread Reply:* Again: two person consensus
*Thread Reply:* “Are metagenomic studies primarily interested in taxonomic abundances and/or presence/absence?”
Depends on the question (and the study system). SedaDNA still uses the latter primarily (due to biases associated with the former).
*Thread Reply:* Hey @Benjamin Vernot, Pete, Irina and I are coauthors of your presentation now right?
*Thread Reply:* at this point, yes!
Is a study that uses metagenomic samples (i.e. sediments), but focuses only on one species / taxa within those samples still a metagenomic study? i.e., are the recent sediment popgen papers metagenomic studies, or do they just use metagenomic samples?
Some examples - are these metagenomic?
Environmental genomics of Late Pleistocene black bears and giant short-faced bears https://www.sciencedirect.com/science/article/pii/S0960982221005406
Genome-scale sequencing and analysis of human, wolf, and bison DNA from 25,000-year-old sediment https://www.sciencedirect.com/science/article/pii/S0960982221008186
Ancient human DNA recovered from a Palaeolithic pendant https://www.nature.com/articles/s41586-023-06035-2
*Thread Reply:* One species, then no. Entire communities: yes.
If more than one species but not a community (the three refs. you list): then good question! I guess pathogen studies also fall within this category, too…
*Thread Reply:* I just chatted with the two pathogens PIs at our institute, and they both said that they don't think of what they do as metagenomics. That they drill into the data too much, to get at the 1-2 species they're really interested in.
*Thread Reply:* Yeah, as in the other thread: apply metagenomic approaches at the intital stages and then genomics afterwards
Hi @Miriam Bravo 👋 👋 👋 👋 👋 👋 👋 👋 (@Shreya told me to say it)
@channel informal demographic poll 👇
Which country are you based in for your work, please leave the country emoji on this post!
HAAM meetup: Let’s meet up in person at ISBA10 in Tartu! We booked the Helmi Kurrik Lecture Hall for a small meet&greet on 14th of September from 10.00-10.30. Pass by with your coffee ☕
RE
https://spaam-community.slack.com/archives/CPYE64ZC6/p1693033331765469
We unfortunately were unable to find a place that could take everyone sorry 😞 but if you want suggests for restuarnts, I'm sure @Meriam Guellil or @Marcel Keller or @Biancamaria Bonucci can suggest places (or see the SPAAM5 whatsapp chat for recommencations from @Biancamaria Bonucci)
Heiiii, gonna write here and in whatsapp to! So there are different places you could go: • Kolm Tilli (different kinds of food, from pizza to baos) • Restaurant München (german food, but germans go there so I imagine it’s good) • La Dolce Vita (Italian restaurant) • Kampus (they have different kinds of foods) • Pompeii (italian-modified food) • Veg Machine (vegan food, but it closes ‘early’, meaning at 21.00) • RP9 (pub dishes) Hope it helps :smilingfacewith3hearts:
*Thread Reply:* you're awesome
*Thread Reply:* Dolce vita is closed this week!
If anyone is not so excited about pop-gen
*Thread Reply:* Thanks! This might actually get me through multiple PCAs 🙌:skintone2:
*Thread Reply:* I just added a bunch of kinship ones too !
*Thread Reply:* How did I do? :)
*Thread Reply:* I only got 2 ;)
*Thread Reply:* So good job 😂
*Thread Reply:* Hah no, I meant on the bingo cards :)
*Thread Reply:* That's what I mean! Oh wait, you think getting a bingo is good?!
*Thread Reply:* gotta keep my popgen cred!
Hello! For the people at #ISBA10. We are having a #spaamtisch de-briefing of #ISBA10. Topic: which were your favorite talks and why? When: from 15 to 15:30 Where:Helmi Kurrik Lecture Hall
PIs join us! We want to have your positive takes on all the students' talks and posters!
See you there!
P.S. the meeting it's very informal!
Thanks to everyone who came to the semi-ninja #spaamtisch 👆
And congrats to @Oya Inanli for winning the hastily-devised and informal inaugural SPAAM Award of the most popular talk of ISBA10!
For another bit of fun, following the success of the poop-gen bingo, we brainstormed a bunch of ancient metagenomic presentation memes/stereotypes/tropes etc to eventually make a ancient metagenomics bingo!
If you have ideas of more cards please join <#C05SX35UMJM|spaam-bingo> and leave a message with your bingo card ideas - the sassier the better!
*Thread Reply:* Many thanks everyone! The beers are on me (in 5 years) 😊
*Thread Reply:* I don't want it if it's not yelled at! 😁😁😁:spaamtisch:
Before and during ISBA10 I talked with members of multiple research groups that work already or plan to work with the „Santa Cruz Reaction“ ssDNA library protocol. It seems a lot of people have problems and already tried to optimize the protocol, or made changes to adapt for their needs. Therefore I created the channel <#C05TER4G9K2|scr-protocol> so we can talk about our problems, experiences, experiments and adjustments of the protocol. Feel free to join!
Hi @channel I hope those of you at ISBA have travelled back safely. In case you just didn't get your fill of metagenomics and need something fun to read our latest SPAAM blog is up on the website. This post gives a great history of SPAAM including where it got it's name: https://www.spaam-community.org/blog/2023/09/11/blog-history/
*Thread Reply:* Omg @James Fellows Yates I just learned the meat thing of the name spaam 🤣 this is brilliant 👏 👌 and huge thanks to you for creating this amazing platform ✨
Hi all! I've seen that the EAA 2024 call for sessions is now open, deadline is the 9th of November!🏛️ https://www.e-a-a.org/EAA2024/Home/EAA2024/Home.aspx?hkey=ca9cb97a-9463-48dc-91b7-b8b214963ad8#Program
@channel Should we propose an ancient DNA metagenomics session at EAA 2024 ? ⬆️
*Thread Reply:* I think we could manage to make it fall under this theme "Archaeological Sciences, Humanities and the Digital era: Bridging the Gaps"
@channel I've created the channel <#C05TBKJ5HU1|eaa-2024-rome> so we can discuss sessions proposals for the ones who are interested!! 🏛️
Any volunteers for an Americas mini-SPAAM/conference session for next year too? Most popular from the SPAAM5 poll as aaba
Hiii Everyone (@channel)! :salute: I'm interested in creating the ultimate overview of all the different metataxonomic classifiers and pipelines out there. I'm already elbows deep into making a sheet with everything in it - for every classifier I want to specify the type of method, it's limitations, give some notes to whether it's annoying to run and/or install etc. Buuut this could take a while to do as a single person so here is where I think it would be cool if some of you wanted to join the initiative. I propose to have a quick zoom meeting about this within the next weeks to chat and brainstorm about how to best implement this. Could anyone interested please react with "👾" and thereafter we could figure out when fits everyone's calendars?
Edit: So happy to see you guys’ interest in this! 🙌 if anyone else wants to tag along then feel free to join the <#C05SQA7RA81|yet-another-taxonomic-profiler> channel! 🧬
Hello from your friendly neighborhood Engagement Officer. A couple new channels for your enjoyment/interest: <#C05U222543A|espaamñol>: for Spanish (SPAAMnish?) speakers! To discuss outreach/training initiatives and potential meetups in Spanish #spaamtv: giving a talk soon? want feedback? post a link to a recording of you rehearsing for people to watch and comment on in their own time <#C05T6CYHYHL|spaam-across-the-pond>: for non-Europe-based SPAAMers to make sure the community is accessible to and serving everyone!
heya all -- the steering committee is working to put together the fall newsletter and wanted to make sure we let people get some input if they want to highlight anything -- think new initiatives, papers, interesting/relevant conferences + courses, job/phd advertisements etc!
-- please reply below if you want anything added and we will try to include it 🙂
*Thread Reply:* Hey Ian, Kadir and I will start the initiative #MetagenomicsMondays where we will tweet papers from the microbial field, old, new and sci-com.
@channel 🎉 New from the SPAAM Blog team (@Shreya and me)! We really mean no stupid questions in <#C02DCKJ54JX|no-stupid-questions> but we get it, sometimes you’re just too nervous to ask! Enter the “I’m too afraid to ask” form where you can post your anonymous questions! Shreya and I will check in periodically and share submitted questions with our community… and maybe make a blog post out of the answers. You can also use the form to anonymously submit blog post ideas. It is pinned at the top of the <#C02DCKJ54JX|no-stupid-questions> channel whenever you need it! 🥳 Have fun!
Hi SPAAM crowd! Does anyone already have experience sequencing (ancient metagenomes from dental calculus) on the new Illumina NovaSeq X Plus instruments with XLEAP-SBS chemistry? We are considering producing at least parts of a dataset on this machine but are not sure what it might mean (just remember how updated chemistry and basecalling approach have been affecting compatibility btw older Illumina instruments). We will also want to integrate the new data with already existing datasets that have been produces on the NovaSeq 6000 series. Any and all thoughts (and experiences) welcome!
What are peoples general approaches to 'reducing' their database sizes for classifiers (whether that's redundancy removal; compression; parameters etc.)
*Thread Reply:* • select limited number of representative genomes for each taxa • if using kraken2/krakenUniq, I would lower the kmer-size to 32/30 to build the db, of course it will increase the chance to get FP on further metagenomic classification
*Thread Reply:* https://github.com/msabrysarhan/taxonize_genbank We wrote this python package to filter the NCBI non-redundant protein or nucleotide databases based on taxonomy so we can confine the search for specific taxa, when using diamond for example (This is in case of dietary analysis). It saves a lot of time 🙂
*Thread Reply:* this is our solution to build our DBs:
Hi all, I would like to promote the Nordic Geological Winter meeting 10-12 January 2024 in Gothenburg, Sweden. There will be a special session on “Sedimentary ancient DNA applications to Quaternary Geology”, organised by @Pete Heintzman, me and some other researchers from Stockholm University.
We are very pleased to have Karina K. Sand as a keynote speaker in our session, and furthermore Inger Greve Alsos is a plenary speaker at the conference, so there will be a lot of aDNA related stuff! We have reserved some talks for ERCs as well, so make sure to submit your abstracts! Abstract deadline has now been prolonged until this Sunday 15th October.
See https://www.ngwm2024.se for more details.
Hope to see you there! 😄
Hi all, there were a lot of people interested in sedaDNA at SPAAM5, might be a lot here as well. Do you guys want to join the <#C01B7CRJK7U|seda-dna> channel? It has been quiet for a long time, maybe we can resurrect it?
Hi all - does anyone have a favorite paper(s) to read as an "intro to ancient metagenomics"? We have a couple new undergraduates starting in the lab and am trying to come up with a basic reading list to give them some context!
*Thread Reply:* this feels like an obvious one, but someone has to mention it! https://www.annualreviews.org/doi/abs/10.1146/annurev-genom-091416-035526?urlver=Z39.88-2003&rfrid=ori:rid:crossref.org&rfrdat=crpub%20%200pubmed|https://www.annualreviews.org/doi/abs/10.1146/annurev-genom-091416-035526?urlver=Z39.8[…]03&rfrid=ori:rid:crossref.org&rfrdat=crpub%20%200pubmed
*Thread Reply:* Of course! Thanks for tossing it out there 🙂 also curating favorite papers involving shotgun seq to have them read occasionally as the semester goes on
*Thread Reply:* Hmmmmm “ancient metagenomics bibliography” seems like a great blog post … I will keep thinking about papers and also keep an eye on this thread!
*Thread Reply:* I think I had a thread of this similar thing to
*Thread Reply:* Maybe more options there too!
*Thread Reply:* These 2 papers may also help.https://pubmed.ncbi.nlm.nih.gov/25487328/,https://pubmed.ncbi.nlm.nih.gov/31638147/


*Thread Reply:* Thank you all for this! This is great.
Hello @channel I hope your summer/conference break was nice! It's Spooky Season and what better way to scare away our metagenomics ghosts if not with a spaamtish séance???? Our first meeting will be in the "regular's table" format. We'll socialise and discuss the future of spaamtisch. But fear not we'll also have a topic to discuss that you'll choose.
For the new people here #spaamtisch is a monthly get together of the spaam community in two formats: discussions and seminars. Go to the #spaamtisch channel for more info and to participate to this initiative!
Feel free to take out your best witch/warlock hat for the occasion:female_mage:
Please go to the #spaamtisch channel and choose a preferred date and time in the poll.
Cheers! The spaamtisch organisers
🚨✨Hey @channel!! We're super excited to share SPAAM Newsletter #2 with all of you. Just click this link and enjoy! :spaam::party_dino:😀🎉https://preview.mailerlite.io/preview/646990/emails/102162115619456606 The sections that we covered are:
- SPAAM Community Initiatives/Calls for help! :manwomangirl_girl:
- Job Positions 💻:femaleconstructionworker::skintone6:
- Upcoming Conferences 💬
- New Publications 📜
- Spread the word for SPAAM-like members in other related fields (HAAM + PAASTA) 🍖 🍝
Enjoy reading and until next time.
Your SPAAM Steering Committee, @Miriam Bravo, @Ian Light, @aidanva, @Maria Lopopolo, @Kadir Toykan Özdoğan, @Betsy Nelson, @Shreya and @Gunnar Neumann
P.S. If you want us to advertise any SPAAM relevant jobs offers, articles, conferences, workshops etc. in our upcoming newsletters, please contact us directly. We try to be up to date as possible and include everything we are aware of, but sometimes even our eagle eyes can miss something.
*Thread Reply:* Awesome thank you Miriam! Just wanted to mention there is an issue with the PAASTA twitter link - it wants to go to @peaas_community instead of @paastacommunity
Can I also selfishly add my own publication to the list? Only if you think it’s appropriate of course! https://onlinelibrary.wiley.com/doi/full/10.1111/1755-0998.13835
*Thread Reply:* Hello Dawn! Thank you for the correction and sharing your paper !! I’ll update the newsletter accordingly! 🙂
Dear @channel we, the spaam community media team, would like to invite you all to help us with the #MetagenomicsMonday initiative. Therefore, we kindly ask you to fill out the form below if you felt particularly inspired by a paper (e.g. a paper that for you was very important in defining your research field) or you think a paper is a milestone for ancient metagenomics, pathogens, sedaDNA, and ancient eDNA. We appreciate everyone for their help!
https://forms.gle/pKVhAeRJCCrQYFh16
Many thanks Maria and @Kadir Toykan Özdoğan
Form is pinned in the channel
Dear all, the people have spoken. Our first #spaamtisch meeting “regular’s table” format will be on the 26th of October 4-5pm CET. The discussed topics chose by the #spaamtisch participants will be:
-Kmer matching vs mapping methods -databases
Additionally the organisers would like to take some time to talk about the future of spaamtisch. In particular we need help in finding volunteers for the seminar sessions who want to present their work. So we’ll talk about proposed strategies to find people 🙂
Many thanks! The spaamtisch organisers
Zoom link will be shared in the next few days.
Dear all! Do you wanna discuss metagenomics, lab issues, job position or anything really in turkish and you don’t know where? The <#C062Q812XNW|spaam-turkish> channel has you covered! We are looking forward to welcome you in the channel!
Dear @channel, find below the link for tomorrow's #spaamtisch meeting!
Biancamaria Bonucci is inviting you to a scheduled Zoom meeting.
Topic: SPAAMtisch Time: Oct 26, 2023 04:00 PM Amsterdam, Berlin, Rome, Stockholm, Vienna
Join Zoom Meeting https://ut-ee.zoom.us/j/95645964400?pwd=ckZqRXNiSTYySEFGazhDOE85d3p3UT09
Meeting ID: 956 4596 4400 Passcode: 278708
@channel we are on zoom for #spaamtisch databases and kmer vs mapping🔥
@channel Hello, hola, merhaba, ciao, et cetera! A quick note from the Steering Committee: We are delighted by the language channels forming (<#C05U222543A|espaamñol>, <#C062Q812XNW|spaam-turkish> and the brand new 🇮🇹🇮🇹#spaamghetti 🇮🇹🇮🇹 so far)! We hope these will be a great resource for local networking, job hunting, or discussing initiatives like educational resources in your language of choice. At the same time, we really hope your voices stay a part of our international general community! We suggest that if an interesting discussion arises in one of the language channels, that someone copy the question in one of the more general channels so a more inclusive conversation can also happen. If you would like to create a community channel, feel free to do so and publicize it! Or ask one of the SPAAM engagement officers (@aidanva and @Shreya) to create and share it for you.
Dear @channel our next #spaamtisch meeting will be in the seminar format. @Jonas Niemann from the GLOBE will present his amazing work on “Parasite detection in ancient and modern metagenomic datasets”. Don’t miss this talk if you are curious about this challenging task!
Please head to the #spaamtisch channel to express your day of preference in the poll 🙂 Cheers, The spaamtisch organisers.
Hello all!
We are excited to announce the SPAAM5 online event of the term, SPAAM5 Part 2!
We hope that everyone who attended SPAAM5 in Tartu had a great time! Now, as promised, the time has come for the SPAAM5 online event, “SPAAM5 Part 2” which will be held on Tuesday and Wednesday, January 30 and 31, 2024 on Zoom. So it is time to submit abstracts! Please submit an abstract of a maximum of 200 words to https://docs.google.com/forms/d/e/1FAIpQLSdIK2du4nfRi1p5aLJSCMFUrG4hgp1JwE2Ne1lgAZiaXYLNRw/viewform?usp=sharing by 11:59pm CET on November 30th. We would like to particularly encourage people who were unable to attend the SPAAM5 in person meeting to submit. This is a great opportunity to hear about amazing new research from young investigators across the globe without any limitations set by travel or budget. We will try to accommodate time zones wherever possible, so that more people can participate, as was the case with the previous online version of SPAAM. Principal investigators are welcome to participate in this edition, but are politely asked to leave sufficient space for early-career researchers during the discussions. If you submit an abstract, please plan for a presentation in the form of a 10-minute talk followed by 3 minutes of questions. There is no registration fee. Thanks very much and we look forward to your submissions!
Welcome to the second part of Standards, Precautions, and Advances in Ancient Metagenomics, edition 5!
Sincerely, SPAAM5 Organizers (Meriam Guellil, Zoé Pochon, Alina Hiß, Alice Lee, Sierra Blunt, and Megan Michel)
*Thread Reply:* Hello, since I recently started my PhD, is it possible to attend without submitting an abstract?
*Thread Reply:* Yes we will open the registration later on. But we need some abstracts first for it to happen! So far, we're still waiting 😅🙏
Hi @channel, is anyone planning on proposing an anciennt pathogen symposium for SMBE next year?
Dear @channel 🎙️ It is decided our next #spaamtisch meeting will be held on the 13th of November 4-5pm CET.
@Jonas Niemann from the GLOBE Institute in Copenhagen will present his amazing work “Parasites Lost: Parasite Detection in Ancient and Modern Metagenomic Datasets”. Don’t miss this talk if you are interested in knowing more about this groundbreaking research exploring the intricate relationship between hosts and their parasites!
♦️The meeting won’t be recorded as it contains unpublished work. Hence, you are encouraged to follow this seminar!
✨Join the #spaamtisch channel to follow all our seminars/discussions on interesting topics chosen by the community every month!
Cheers, The spaamtisch organisers.
ps Link to be shared at later stage
Hello all,
As Zoé mentioned last week, the organization of SPAAM5 Part 2 is officially underway! This event will be held virtually in order to accommodate the entire SPAAM community. If you attended the in-person meeting in September, we would love your feedback. What did or didn't work for you? What would you like to see at our next meeting? Please share your thoughts with us by filling out this anonymous survey before Friday, November 17th: https://forms.gle/QMF2fvphn5QBc8QC8
Thank you! SPAAM5 Organizers (Alice, Alina, Megan, Meriam, Sierra, and Zoé)
Dear @channel the zoom link for today #spaamtisch at 4pm CET is:
https://ut-ee.zoom.us/j/95645964400?pwd=ckZqRXNiSTYySEFGazhDOE85d3p3UT09
We are connecting, come along if you’d like to hear @Jonas Niemann talking about ancient parasites 🙂
Dear all!! here you can find the form to sign up for presenting at #spaamtisch !!
We would like to point out that we are offering to host more types of presentation. In the form, you will be asked what kind of presentation you would like to give, such as:
• Regular seminar ( to present your newest paper, published or on biorxiv) • Work-in-progress seminar (to present your data, in order to get feedbacks and help with things you are stuck on) • Journal Club (discussion of a paper of your choice) Don’t be shy 🥳 🥳
Cheers, The spaamtisch organisers
Hi @channel! We need some abstracts for SPAAM5 Part 2 (online) :spaam: to happen! Ancient metagenomicists from all over the world, this is your occasion to shine and present your research 🦠✨! Please submit an abstract of max 200 words to https://docs.google.com/forms/d/e/1FAIpQLSdIK2du4nfRi1p5aLJSCMFUrG4hgp1JwE2Ne1lgAZiaXYLNRw/viewform?usp=sharing by November 30th. We will try to accommodate time zones as much as possible, as was the case for SPAAM4.
Hi @channel! Deadline : this Thursday November 30th! We need some abstracts for SPAAM5 Part 2 (online) :spaam: to happen! Ancient metagenomicists from all over the world, this is your occasion to shine and present your research 🦠✨! Please submit an abstract of max 200 words to https://docs.google.com/forms/d/e/1FAIpQLSdIK2du4nfRi1p5aLJSCMFUrG4hgp1JwE2Ne1lgAZiaXYLNRw/viewform?usp=sharing by November 30th. We will try to accommodate time zones as much as possible, as was the case for SPAAM4.
*Thread Reply:* I've got some new sedaDNA work (modern!) that I just got data on (time series, MAGs, etc), but want to leave space for more on topic things. I'm going to submit an abstract, and would love to attend, but feel free to disregard if there's more relevant topics!
Hi All! Just over two weeks left to submit an abstract to the UK Archaeological Science conferences which will be held in York in April 2024. The deadline is the 18th of December, with registration opening at the end of the year. You don't need to be based in the UK to attend. https://conference-service.com/UKAS-2024/xpage.html?xpage=314&lang=en|https://conference-service.com/UKAS-2024/xpage.html?xpage=314&lang=en. PS - York is lovely in the Spring! 🌞
Hey @channel! We extended the deadline for abstracts to next Thursday December 7th! We need some abstracts for SPAAM5 Part 2 (online) :spaam: to happen! 🙏 Please submit an abstract to https://docs.google.com/forms/d/e/1FAIpQLSdIK2du4nfRi1p5aLJSCMFUrG4hgp1JwE2Ne1lgAZiaXYLNRw/viewform?usp=sharing
Dear @channel
We are delighted to let you know that for our #spaamtisch meeting before the season holidays, Santa is bringing us @Nikolay Oskolkov who will be presenting the work on the aMeta pipeline. It will be the perfect meeting for those of you who are interested in pathogen discovery.
Please reply to the poll on the #spaamtisch channel to choose your preferred date✨ Cheers, The spaamtisch organisers
*Thread Reply:* Dear SPAAM organizers, please note that it says "SPAAM5" everywhere in the registration / abstract submission form instead of "SPAAM6" 🙂
*Thread Reply:* Do you mean the online meeting in January? That still belongs to SPAAM5 😉
*Thread Reply:* Aha, I mixed them up, now I see
Hello, @channel save the date for our last #spaamtisch meeting of 2023 on the 15th of December 4-5pm CET !
For all the pathogens and pipelines aficionados, we’ll close this year’s seminar series with @Nikolay Oskolkov who will be presenting the aMeta paper!
Also, I remind all of you that you can sign yourself up https://docs.google.com/forms/d/e/1FAIpQLSc0Lb3kKm2sRTutY_9Nyk58SN9Lg82VfUMtVvrEm_VGmwHZxw/viewform?usp=sf_link and present a work you have recently published, a paper you want to discuss, or a work in progress you need feedback on.
We are looking forward to seeing you soon! Happy weekend, The spaamtisch organisers.

*Thread Reply:* Thanks for sharing that @Barbara! Ngl the link confused me a lot at first 🤣
*Thread Reply:* Congratulations, @Miriam Bravo !!!
Wait whhhaaaaaattt???? Congrats to @Miriam Bravo!!!!!
Translated summary:
This year the 25th edition of the L'Oréal-UNESCO International Award for women in science was held, in which 8 scientists of excellence were recognized through 5 scholarships and 3 awards awarded by the L'Oréal Group in conjunction with the UNESCO, the Mexican Academy of Sciences and Conalmex.
The ceremony, which was conducted by Adela Micha, had as winners Dr. Rossana Arroyo Verástegui, Dr. Erika Bustos Bustos, Dr. Sandra Elizabeth Rodil Posada, Dr. Miriam Jetzabel Bravo López, Dr. Monserrat Llaguno Munive, Dr. Azalia Avila Nava , Dr. Mariana Felisa Ballesteros Escamilla and Dr. Maricarmen Iñiguez Moreno, who received their award from the hands of Ana Laura Rendón, Marion Reimers, Pamela Cerdeira and Deborah Armstrong.
Hello, @channel here is the zoom link for today #spaamtisch at 4pm CET:
https://ut-ee.zoom.us/j/93577052785?pwd=ekxNM2s5NFkzMjhsNUxYa2c2cFFQUT09
*Thread Reply:* will todays spaamtisch be recorded?
*Thread Reply:* Yes!
Hi @channel we are connected! Nikolay will start in 5 min!
@channel we heard you! Please wait for news in 2024 👀 The format changes and so does the date. :meow_party:
Dear @channel 🚨EAA 2024 Calls for Abstracts OPEN NOW! Showcase your groundbreaking work, join our session “Building bridges: An open forum for Archaeology and Metagenomics” where experts from Archaeology and Metagenomics can unite to showcase groundbreaking projects! How : Tandem or solo presentations ⏰Abstracts due Feb 8th (oral) or Apr 8th (poster) 💻 Can’t make it to Rome? No worries! EAA offers online participation. 👉 Submit at submissions.e-a-a.org/eaa2024 with session number 854 More info https://www.e-a-a.org/EAA2024 #EAA2024
Join us in Rome for the EAA 2024 session #704 on ancient pathogens! 🦠Title: Bioarchaeology of Ancient Pathogens 🇮🇹🍝Where: In Rome! 📆When: 28-31 August 2024 ⏰Abstracts due by February 8th!
Interested in the impact of ancient pathogens on human populations, their domesticated animals, and crops? 🐐🚶🌽 Please submit an abstract in our session https://submissions.e-a-a.org/eaa2024/ Feel free to reach out to us, @Zoé Pochon, @Louis L'Hôte, @Rémi Barbieri, Shevan Wilkin @Kerttu Majander! @channel
Dear @channel happy new year! I hope everyone had a nice season holiday and now you all come back with renewed energy for a year of #spaamtisch. Don’t forget that we have a form where you can sign up to volunteer to give a seminar in three different formats: • Regular seminar (where you present a recently published work) • Work in Progress (where you present something you are working on and you need feedback on from the community) • Journal Club (you choose a paper that you are interested in exploring with the community) https://docs.google.com/forms/d/e/1FAIpQLSc0Lb3kKm2sRTutY_9Nyk58SN9Lg82VfUMtVvrEm_VGmwHZxw/viewform?usp=sf_link Go to the #spaamtisch channel to choose a date for the meeting. Don’t forget to add a topic you would like to discuss in our rota of topics document. https://docs.google.com/document/d/1YMcDb5sJMAZeThgKkNea7X5V7pa1eaQx/edit?usp=sharing&ouid=118094628411852884592&rtpof=true&sd=true
Cheers, The spaamtisch organisers
We are pleased to announce session #653 “Infectious Disease and Hygiene in the Neolithic and Bronze Age” taking place at the Annual Meeting of the European Association of Archaeologists (EAA), August 28-31, 2024, in Rome, Italy.
The session aims to create a platform for discussing the interactions between humans and infectious disease during the Neolithic and Bronze Age from a biosocial perspective. To enhance our overall understanding of pathogen presence, diversity and evolution, as well as human responses to infectious disease during prehistory, we welcome contributions from diverse research fields, including paleopathology, bioarcheology, history and genomics.
If you would like to participate with an oral presentation we kindly invite you to submit your abstract following the instructions on the conference website: https://www.e-a-a.org/eaa2024
Please note that the deadline for abstract submissions is February 8th.
For more details on the session #653, please see the abstract below.
Do not hesitate to contact us in case you have any questions.
Best wishes,
Maria Spyrou and Agnė Čivilytė
Abstract: Understanding the interlinked histories of humans and infectious disease promises a more comprehensive view of our past. The investigation of infectious diseases in archaeological contexts has gained increasing interest given the numerous open questions with regard to the aetiological agents of past pandemics and the potential influence of human subsistence and sociocultural changes (such as the Neolithic and Bronze Age transitions) on population health. Throughout human history, a number of epidemics and pandemics have been historically recorded or are hypothesized to have occurred, however, a deep understanding of the influence of infectious disease on human societies prior to the availability of textual records is still lacking. In this session, we aim to use a multi-faceted approach that enhances our overall understanding of pathogen presence, diversity and evolution, as well as human responses to infectious disease during prehistory, with special focus on the Neolithic and Bronze Age periods. To create a platform for discussing the interactions between humans and pathogens from a biosocial perspective, we welcome contributions from researchers specialized in paleopathology, bioarcheology, history, and genomics. Specifically, this session will focus on:
- The identification of infectious diseases in the Neolithic and Bronze Age using a variety of methodological approaches;
- The determination of drivers of disease emergence and transmission, such as living circumstances, dietary habits, food preparation methods, waste disposal practices, etc.;
- The exploration of evidence of hygienic treatment and prevention of disease in prehistoric communities;
- The impact of infectious diseases on the demographic and social development of Neolithic and Bronze Age societies; and
- The characterization of long-term evolutionary history of pathogens and their association to modern-day diversity, especially for infectious diseases that continue to pose a threat to public health today.
Hello @channel
Abstracts are open for the EAA in 2024. If you're into old poop, we have the session for you! 💩
Session 1061: Appreciating the Archaeological Value of Preserved Faeces Using Multifaceted Methods
For more info, including the full session description, please see the attached flyer and let me know if you have any questions.
It would be great to see some SPAAMers there, so @ your fave coprolite / palaeofaeces / archaeological dung folks... Oh and it's in Rome ☀️
*Thread Reply:* Hello, I'm wondering what is the poster presentation DL? On the website the poster presentation deadline is 8th of April https://www.e-a-a.org/EAA2024/GeneralInfo.aspx?WebsiteKey=20b5538d-68f8-4056-9596-1ae1ce0ead47&hkey=d1efbf02-cbd8-4e90-87d9-3ebffb323fc5&NewContentCollectionOrganizerCommon=2#NewContentCollectionOrganizerCommon|https://www.e-a-a.org/EAA2024/GeneralInfo.aspx?WebsiteKey=20b5538d-68f8-4056-9596-[…]-4e90-87d9-3ebffb323fc5&New_ContentCollectionOrganizerCommon=2
Hello I would like to invite you to SMBE-July 7-11 2024 in Puerto Vallarta Mexico:meow_party: It will be a nice a opportunity to share and see each other in person. My colleague Sur Paredes and me (Barbara Moguel) organized the session "Molecular evolution through metagenomics" (https://smbe2024.org/) and our confirmed invited speakers are Nandita Garud (UCLA), and Gabriela Olmedo (Cinvestav). Abstracts are going to be due sometime in February.
Below I copy the description of the symposium which will be published in the SMBE 2024 meeting website. I hope you consider attending:
“The ecosystems across the globe are undergoing significant transformations, underscoring the imperative need to comprehend their eco-evolutionary mechanisms. Until relatively recently, most of the work on molecular evolution focused on the analysis of single species genomes. Recently, metagenomics has emerged as a valuable approach to ask global questions about biodiversity, ecology and evolution at the community level. With metagenomics, we can ask questions about comparative micro- and macro evolution, within and between environments, as well as removing the limits on difficult to culture organisms.
This session offers a unique opportunity to showcase the extensive and consequential methods and applications, both theoretical and experimental, that harness metagenomic data to ask questions about molecular evolution within diverse ecosystems. These ecosystems encompass animal and plant bodies, but also extreme natural habitats, historical landscapes, areas endangered by anthropogenic activities, and more. This symposium will highlight contributions that go beyond the acquisition of taxonomic and functional profiles, and that instead aim to disentangle the mechanistic details of the evolutionary process, focusing either on single species or community assemblies. This symposium aims to highlight the breadth of biodiversity that can be interrogated through metagenomics, which include both the complete extant tree of life, as well as ancient biodiversity which is increasingly important to comprehend the threats of climate change. This symposium will serve as a platform for sharing cutting-edge research, fostering collaboration, and advancing our comprehension of the molecular underpinnings that have sculpted the tree of life.”
For all those adveritsing their various conference sessions:
Don't forget to also advertise on the following (e)mailing lists:
• SPAAM: spaam-community@listserv.dfn.de • ISBA: isbarch@jiscmail.ac.uk • ANCIENT-DNA : ancient-dna@jiscmail.ac.uk :)
*Thread Reply:* And if someone has sessions where palaeoproteomics fit, feel free to send them to me and I'll pass them on to PAASTA (we don't have an emailing list yet) 😊
*Thread Reply:* Get a mailing list!
*Thread Reply:* Sssscchhh we're working on it!
*Thread Reply:* Also welcome back to spaam 😏
*Thread Reply:* I never left. Just turned into a lurker.
Dear @channel 🥳
I would also like to advertise our session at the EAA 2024 :meow_party: :
Session 122: Indicators of Nonprivileged Populations from Funerary Contexts: Multidisciplinary Approaches to Assessing Disparities in Past Populations
Keywords: Inequality, dietary stable isotopes, paleogenomics, paleopathology, funerary archaeology, bioarchaeology
We are hoping for a very diverse session full of interdisciplinary contributions, so please don't feel shy and send us some abstracts :party_dino: If you have any questions, feel free to contact me. Deadline for the abstract submission is: 8. 2. 2024.
Hei @channel, Here the link for today's #spaamtisch at 4pm CET ☺️
Join Zoom Meeting https://ut-ee.zoom.us/j/93577052785?pwd=ekxNM2s5NFkzMjhsNUxYa2c2cFFQUT09
Meeting ID: 935 7705 2785 Passcode: spaam
Vibe check - please vote on the following: "I am getting too many notifications from SPAAM"
:spaam:📣 @channel!! SPAAM Newsletter #3 :spaam:📣
📣 Hello from the SPAAM Steering Committee!
We have collected for you a selection of the available conferences for this year, including info on the relevant sessions, dates, and submission deadlines for abstracts!
Additionally, below we have a reminder/call for submissions to participate more actively within SPAAMtisch, a monthly meeting of SPAAM members to discuss various topics in a low-stress environment focused on learning!
🚨The large sections are:
- Upcoming conferences
- Community initiatives
- Open job positions
- Recent publications
View SPAAM Newsletter #3 ----> https://preview.mailerlite.io/preview/646990/emails/112035890131371356
Enjoy reading and until next time. Your SPAAM Steering Committee @Miriam Bravo, @Ian Light, @aidanva, @Maria Lopopolo, @Kadir Toykan Özdoğan, @Betsy Nelson, @Shreya and @Gunnar Neumann
*Thread Reply:* Very helpful, thanks a lot!
Dear @channel February is here and I am very pleased to tell you that this month for #spaamtisch our @irinavelsko will present her Biorxiv preprint on dental calculus from the Pacific!
Please go to the #spaamtisch channel and choose your preferred date for the meeting🙂 We will also record the meeting and prepare a similar Google doc for continued questions from the seminar. Cheers!
Hello everyone 👋
We are planning our third edition for the SPAAM summer school - introduction to ancient metagenomics!
As always, I would like to ask if anyone would be interested/willing to be a *teacher* or an *instructor* for any of the sessions. The material we teach can be found in the companion Textbook.
As a teacher, you get experience teaching (obviously), official certificates from Friedrich-Schiller-Universität Jena and Harvard University as proof of instruction, and also potentially co-author in the textbook (if you add/update content) if we push to publish it 'officially'.
Please note we are looking mainly for advanced PhD students and/or PostDoc level researchers
If you're interested, please DM me or @Christina Warinner for more info 🙂
(in particular if there is anyone working in sedaDNA, we would like to improve the content in that area)
Just saw this from @Claudio Ottoni: https://paleogenomics-tor-vergata.readthedocs.io/en/latest/index.html
Looks like lots of useful training material!
*Thread Reply:* Thanks @James Fellows Yates! We use it as general reference for master students to practice indeed. Not really up-to-date in some parts, but I promised myself to refresh it soon. Tips/comments/corrections are welcome! Hope this helps!
*Thread Reply:* Great to have everything open 😄 💪
Hello @channel! The SPAAM6 organizing team is looking for people from Univeristá La Sapienza (Rome) that are planning to attend the SPAAM6 meeting and are willing to join the organizing team!
Hi @channel -- This is Ian, the SPAAM secretary. I am here to ask you to fill in a very (very very) short form which will help us fulfill our reporting obligations to ISBA. The form can be completed in <1minute and collects basic information about which institutes we represent. Your name and contact details will not be collected with the form.
Please take a moment to fill it out when you can! Thanks so much!
https://forms.gle/kFTPJBkvEHqP72oM7
🎉 🦠 :flag_mx:
@channel Abstract submissions are OPEN for SMBE 2024 and due March 15! Submit here: https://smbe2024registration.org/abstracts We will have a symposium called “Unveiling the evolutionary history of pathogens through paleogenomics” with invited speakers Dr. Tanvi Honap and Dr. Elizabeth Nelson. SMBE 2024 will be in Puerto Vallarta, Mexico from July 7-11, 2024. More info: https://smbe2024.org/
¡Nos vemos en Puerto Vallarta! @Miriam Bravo, @Gunnar Neumann, @aidanva, and Shreya 🧬 🦷 🏝️
Hello everyone #spaamtisch with @irinavelsko presenting is going live now! Join us! Join Zoom Meeting https://ut-ee.zoom.us/j/93577052785?pwd=ekxNM2s5NFkzMjhsNUxYa2c2cFFQUT09
Meeting ID: 935 7705 2785 Passcode: spaam
Dear all,
Tech Tuesday at the American Museum of Natural History (AMNH) These monthly meetings hosted by the Institute of Comparative Genomics (ICG) department at the AMNH are dedicated to talks on new and exciting techniques using genomic data (either from modern or historic/ancient tissues). For the next edition of Tech Tuesday, we’re pleased to announce that we will be joined by Dr. Steven Fiddaman for a talk on the evolutionary history of Marek’s disease virus using ancient DNA techniques next
Tuesday, February 20th from noon-1pm ET over Zoom at the link below. Zoom link: https://amnh.zoom.us/j/91328829642?pwd=d3NlU2lSaFYxUjlIdG5jT0tkamV0dz09&from=addon
Dr. Fiddaman is a post-doctoral researcher at the University of Oxford where he studies everything from rapid (and recent) evolution associated with the domestication of farm animals to adaptation associated with penguins living in different climates. Steven’s talk at the AMNH Tech Tuesday will focus on his recent publication in Science (which you can find here) on the evolution of virulence in Marek’s disease virus (MDV) in chickens. Over the past century, MDV has seen a marked increase in virulence and today, MDV infections kill >90% of unvaccinated birds. In their 2023 publication, Fiddaman and team leveraged ancient DNA techniques to sequence MDV genomes from archaeological chickens and demonstrated that the disease has been circulating for at least 1000 years. This work highlights the growing field and utility of functional paleogenomics and how ancient DNA may be used to understand the underpinnings of pathogen virulence.
Hi @channel -- Just another ping about the demographics form -- it really takes only 30 seconds! Thanks to everyone who has already filled it out. It is important that we get as close to the entire community as possible!
Please take a moment to fill it out when you can! Thanks so much!
https://forms.gle/kFTPJBkvEHqP72oM7
Dear all, I am pleased to announce that @Giulia Zampirolo from GLOBE Institute will present her work on “Early Pastoralism in Central European Forests: Insights from Ancient Environmental Genomics” for our next #spaamtisch meeting.
Please vote your preferred date in the poll I created in the #spaamtisch channel. Cheers, The spaamtisch organisers
Dear @channel,
I would like to raise your awareness about a meeting related to our first community project, AncientMetagenomeDir (found here #ancientmetagenomedir).
The project is up and running since July 2020 and has served as a great example of how the engagement of many members of the SPAAM community across multiple institutes can lead to great resource for information for the whole field of ancient DNA. Its companion publication (https://www.nature.com/articles/s41597-021-00816-y) has been cited almost 20 times in the three years since its publication.
However, we, the organisational team of AMDir, realised in the last couple of months a decrease in the community participation in the project. We are aware that this is a common phenomenon of any voluntarily run project but we would like to change this. 🙂 @James Fellows Yates and I have brainstormed some ideas that we would like to discuss with everyone who would be interesting in contributing to this resource in the future.
If you are interested to hear and discuss about the next steps in the AMDir development, please fill out this when2meet poll and mark your availability for such a meeting: https://www.when2meet.com/?24332943-5AThg In case you are interested, but cannot make any of the proposed dates, please let me know directly.
I am looking forward to discussing the future of AMDir with many of you!
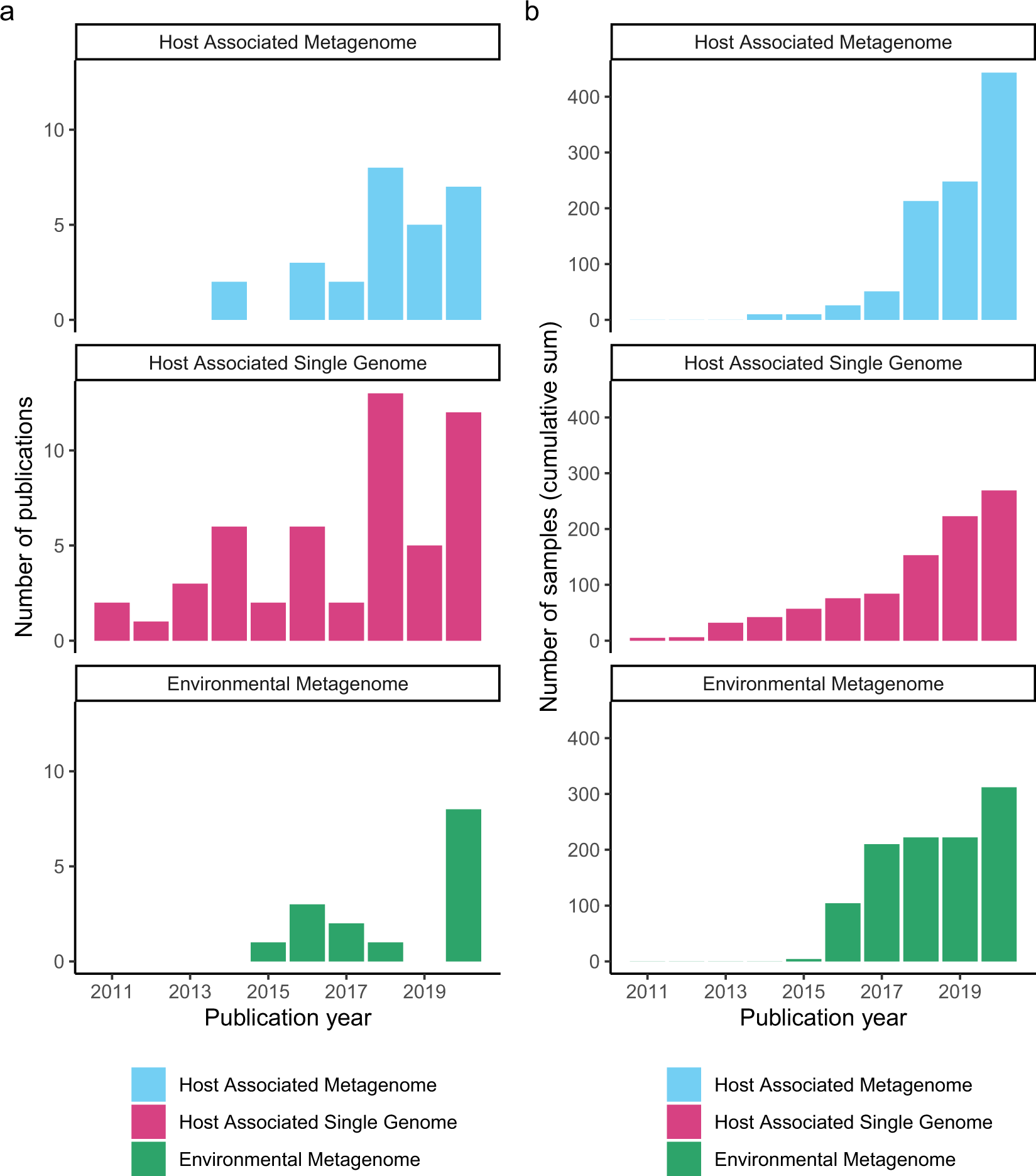
@channel Please note with the above: if anyone from APAC (Australia, NZ, etc.) are interested as well, please let us know - when2meet times are set based on the majority previous contributors timezones, but all are welcome so we can find a different solution in that case!
Hello everyone #spaamtisch with @Giulia Zampirolo presenting is going live in 13 minutes! Join us!
Join Zoom Meeting https://ut-ee.zoom.us/j/93577052785?pwd=ekxNM2s5NFkzMjhsNUxYa2c2cFFQUT09
Meeting ID: 935 7705 2785 Passcode: spaam
And it's that time of the year again! The third SPAAM Summer School: Introduction to Ancient Metagenomics is now open for applications!
It will again be hybrid (with focus on online gather.town, in-person in Leipzig at own cost), and *will be running 5th to 9th of August 2024*
Get more info here: https://www.spaam-community.org/wss-summer-school/#/
*Applications shut on May 31st*
*Thread Reply:* Hi James - I wonder what's the cost associated with the online summer school? Will need 💰 support from my supervisor! :party_dino:
*Thread Reply:* 0 cost, it's free!
*Thread Reply:* @James Fellows Yates Thanks!!!! whoo whoo
*Thread Reply:* Hi, damn I missed the application deadline 😔 I would be super interested to join online, does it make sense to send application anymore?
*Thread Reply:* You've not missed it, it's open until 31st of may :)
*Thread Reply:* Applications are still open!
The focus is online, just if you want to also be physically in the room in Leipzig you're welcome to do so but must pay/organise for your own flights and accomodation etc.
But it's free to join online, if you get accepted
Hi everyone,
This is just a short reminder for the topic of discussing the future of AncientMetagenomeDir: https://spaam-community.slack.com/archives/CPYE64ZC6/p1711540972353019 If you haven't added your availability yet, please do so here (https://www.when2meet.com/?24332943-5AThg) by tomorrow evening. I will close the poll and announce the data on Friday, Apr 5th.
Alright, thank you all for participating in the poll. Unfortunately, we weren't able to find a date and time that would fit all. The greatest overlap with everyone that was Friday April 12th at 2 pm CET. I would aim for the meeting to last approx. an hour, give or take. I will send a Zoom link closer to the date.
*Thread Reply:* Wait so it's 2 pm CEST? I was confused when the meeting said, starts in 1h 😅
As promised, here is the Zoom link for the meeting on Friday:
https://eu02web.zoom-x.de/j/62830727319?pwd=TmdmcW5IdFpIZ1ZjeGJuOUpKeW5hdz09 Meeting ID: 628 3072 7319 Passcode: 328198
I just want to reiterate that everyone, even those who didn't fill in the initial survey, can participate in the meeting on Friday. I am looking forward to seeing you all then!
@channel future of ancient metagenome dir meeting zoom link is now open!
Hi @channel! The SPAAM5 committee is planning an online FAQ session for you to meet experts in different topics of the field of ancient metagenomics. Please sign up, if you are interested in a personal one-on-one discussion with experts in your field to discuss questions which might have come up during your work.
Registration link: https://forms.gle/7CBh5R4Eqhk1dYAJ6
Date of the FAQ session: June 10th from 2 to 6pm CEST (Note that some experts will be available at a different date, preliminary June 14th)
Wetlab protocols - Rodrigo Barquera nf-core/eager - James Fellows Yates aMeta - Zoé Pochon Microbiome classification/analysis - Irina Velsko Pathogens - Meriam Guellil De novo assembly - Alexander Hübner sedaDNA - Peter D. Heintzman Environmental microbes - Antonio Fernandez Guerra
Hi @channel, In our last meeting, we have decided that it would make sense to have a dedicated AncientMetagenomeDir core team to ensure the successful future of AncientMetagenomeDir. In case you are interest to learn more about AncientMetagenomeDir or even want to join the core team, please check out the #ancientmetagenomedir channel for more information.
@channel To continue with SPAAM project related events 😄
*SAVE THE DATE: MInAS ancient DNA metadata standards development hybrid workshop on 13th or 14th of June (provisional)*
We are happy to announce a one-day MInAS Development Event will be provisionally held 13th or 14th June 2024 (the exact day to be confirmed very soon) between 09:00_16:00 CEST.
The MInAS project aims to develop standardised metadata reporting schemes of ancient DNA samples and sequencing data via community-based consensus and training.
This one-day online/hybrid event will involve:
• Introductory presentations of the project • Development of the next version of the MInAS checklist (v0.2.0) • Planning and strategy discussion of the project • Training on how to give checklist feedback sessions to other groups / reading MIxS or MInAS checklists • Generation of additional discipline-specific ancient DNA metadata checklists The event will be online using Zoom or gather.town, and for those who can make it in-person in Copenhagen, Denmark.
If you're interested in joining, please email info@mixs-minas.org with:
• your name • preferred email address • affiliation • main research focus to register your interest and get updates.
Kind Regards,
James (on behalf of the MInAS Team)
@channel Happy Monday Everyone!! ☀️
If you have a spare twenty minutes today to help with one of our community projects, please join us over here on gather! We will be there from now and for the rest of the day! Even 10min could do:)
@channel please sign up if you are interested in joining! Deadline is the May 6th 🌟
Dear @channel On our next #spaamtisch we will have the MInAS Team asking YOUR perspectives and contributions to making a field-wide reporting standard! They will introduce the project and ask you to try filling out the draft MInAS metadata checklist they have been developing. 🗒️💻 If you want to provide input, contribute and make sure the standards fit your research, join us on May 6th at 14:30 CEST!
More detailed information and the zoom link can be found in the #spaamtisch channel!!! 👈
Cheers, The SPAAMTisch organisers and the MInAS Team
Hi all!
Just a reminder you have less than a month to apply for this year's SPAAM summer school!
https://spaam-community.slack.com/archives/CPYE64ZC6/p1712050603518269
Cheers, Ja,es
And @channel please come and join #spaamtisch starting in TEN MINUTES!
It requires your *active participation* to contribute to a critical research infrastructure! And contributions can possible result in coauthorship 😄 (if you need something to sweeten the deal 🍰 )
Oh and I have stickers
If people want more material bribery
@channel Deadline extended to May 9th! Please sign up if you are interested in joining! 🌟
Hi @channel! I am happy to announce the first Cross-Community event (SPAAM, HAAM, PASTA and AaRC) on the 16th of May at 10:00 CEST The Webinar will consist of a presentation by @James Fellows Yates and a Q&A roundtable
Title: Improving reporting and reproducibility of lab work in biomolecular archaeology: saving your, your colleagues, and future you’s time! ```Reproducibility in science has emerged in recent years as a hot topic within all related disciplines. Internet-based open-source software and reproducible notebooks have greatly facilitated transparency and reliability in scientific results during data analysis components of projects. In contrast, similar developments at the lab work stage of a project have been much slower.
The experience of the reporting lab methods of myself and many of my colleagues in a variety of fields is that of a tedious, boring, and time consuming process. This has resulted in common poorly-described methods sections and non-reproducible protocols in many studies, impeding the ability of other researchers (and even ourselves) to adopt new state-of-the-art protocols, or understand how public data was generated.
In this talk I will introduce how lab methods and protocols reporting can be much simpler, accessible, and ultimately save large amounts of researcher's time through the use of citable protocol sharing platforms. I will demonstrate this using the platform https://protocols.io as an example. By the end of the talk and the following Q and A session, I hope to have convinced you to think differently about lab method reporting and provide possible tools to facilitate this critical part of the scientific process.``` A flyer with the registration link will follow shortly!
Here is the flyer! You can also just register here!!!
📣 :spaam:Hello from the SPAAM Steering Committee! :spaam:@channel!
:partydino:Enjoy reading the SPAAM Newsletter #4 and until next time ---> access through this link👉:skintone_6: https://preview.mailerlite.io/preview/646990/emails/120826521034163946
The large sections are:
- Announcements 🔈
- Upcoming conferences ✈️
- Community initiatives 🤝
- Open job positions :catjam:
- Recent publications 📜
P.S. If you want us to advertise any SPAAM relevant jobs offers, articles, conferences, workshops etc. in our upcoming newsletters, please contact us directly.
Your SPAAM Steering Committee Miriam, @Ian Light, @aidanva, @Maria Lopopolo, @Kadir Toykan Özdoğan, @Betsy Nelson, @Shreya and @Gunnar Neumann.

:spaam: Hello @channel from the SPAAM6 organising committee! :spaam:
We are super happy to announce that the conference you were all waiting for is here! :partyparrot:
• SPAAM6 is happening in Rome this year, in the same venue that EAA on the 28th of August. 🇮🇹 • We are also planning to have the possibility to attend online in case you can’t make it to Rome. 🧑💻 And and and we would love to hear about all of your work/thoughts/plans! Hereby we welcome all who would like to present their work on metagenomes, ancient pathogens, methods to analyse metagenomics or microbial genomics datasets, etc. to submit an abstract.
To submit an abstract please fill out the registration form here!
We are looking forward to reading your cool research and remember that the abstract deadline is on the 24th of June 2024 at 23:59 CET!
Cheers, @Ele, @Martin Nathan, @Louis L'Hôte, @Davide Bozzi and @Aleksandra Laura Pach
@channel For those who registered for the cross-community webinar, we are starting in 10 minutes! Feel free to start joining the Zoom link already you should've got from Victoria Mullin (AaRC)
https://spaam-community.slack.com/archives/CPYE64ZC6/p1715157331103869

*Thread Reply:* Definitely posted that in the wrong slack chat!!
*Thread Reply:* Tee-hee, I thought so 😬
*Thread Reply:* It's a cool idea though! Maybe @Miriam Bravo should set this up for spaam, like profiling members of the community to help everyone get to know each other a bit more
*Thread Reply:* Sure ! Sounds good , I’ll look more into that !
Hi guys, does anyone know who is currently leading the aDNA lab in Zurich?
*Thread Reply:* My wild guess was that it was the mummies group, at least from the university website, but I'm also wondering
Hi everyone!
I just want to introduce to you the latest member of the <#C05F0URFJ7M|little-book-smiley-plots> family! Created by the wonderful @AK Runge and @Petra Korlevic!
Meet Sebastian Sprudelwasster, and Sasha! Who represent weird damag epatterns from proof-reading enzymes, and insufficient reads respectively
If you're interested in contributing either your own weird damage plots, but also in particular little characters (quality is not important!) please head over to <#C05F0URFJ7M|little-book-smiley-plots> and let us know and we can give you instructions on what to do :)
i love how they are polar opposites in music enjoyment. 🎶 one is loving the rave the other is having NONE of it
Reminder that the summer school applications close end of this week!!
Hi @channel! We are very excited to announce a new event: Speed networking between PIs and young researchers! We will group PIs and young researchers into pairs/small groups in breakout rooms to share a bit about their background, all within a limited amount of time. We have planned this event to replicate the informal networking that happens in a conference but from the comfort of your own house 😉
The first date for this event series would be the 17th of July at 16:00 CEST (Paris time)! Come to talk with: • Prof. Anne Stone (Arizona State University, https://stone.lab.asu.edu/). Topics: ancient pathogens, mycobacteria, primate variation, human population history, forensics. • Associate Prof. Dr. Christina Warinner (Ph.D.) (Harvard University, Max Planck Institute for Evolutionary Anthropology, https://christinawarinner.com/ ). Topics: oral microbiome, gut microbiome, food culture through microbes, proteomics, human adaptation to extreme environments • Dr. Nicolás Rascovan (Institut Pasteur, https://research.pasteur.fr/en/team/microbial-paleogenomics/). Topics: oral microbiome, ancient pathogens, human population genetics, evolution • Dr. Katerina Guschanski (The University of Edinburgh, Uppsala University, https://www.uu.se/en/contact-and-organisation/staff?query=N14-1484). Topics: microbiome, evolution, ecology, nonhuman primates, biological diversity This is your chance to get to talk to them in an informal setting, to ask them any questions and see if they may be a good fit for your future career! If you are interested in participating, please fill out the registration form by July 10th.
Please note -- slots are going to be given on a first-come-first serve basis! so please sign up soon if you are interested!
Hi @channel ! SPAAM6 abstract submission deadline has been extended to July 7th 23:59 CET! SPAAM6 will be held in Rome this year, in the same venue of EAA, on the 28th of August.
To submit an abstract please fill out the registration form here: https://docs.google.com/forms/d/e/1FAIpQLScwPrmJwS5mdOj9SmCvSdDngTo0H-tx5Mq5b7cgB6C_7MGlcQ/viewform
For organization purposes, if you are not planning to present but you are still interested in attending react with this emoji to this message: :partyparrot:
And if you would like to join remotely (presenting or attending) please answer with the following emoji: :catjam:
Cheers! On behalf of the SPAAM6 organizing team
Hi everyone! We are pleased to announce that the next International Symposium on Biomolecular Archaeology will be held in Turin, Italy, from the 26th to the 29th of August 2025. Stay tuned for updates by following our social media channels: Instagram: @isba11turin X: @ISBA11Torino Website: www.isba11.com We look forward to welcoming many of you next summer!
@channel I'm writing this as a part of my role of ISBA webmaster (the society that spaam is affiliated under).
If you hadn't already heard, Slack is changing their policy to not just hide all old messages after 90 days, but they will from August will delete all messages older than 90 days.
This is really really bad for us in terms of the slack being a useful knowledge source.
Therefore the ISBA society is currently considering moving to a matrix /element server (https://element.io/). This is a much cheaper alternative and means we retain control over our data (can't be locked behind a pay wall). One downside is that we have to maintain this ourselves (i.e. stability will not be guaranteed in the same way as a large and rich commercial company can).
I would like to ask for your opinion again as this has now become more pressing.
I will an (anon) poll below i would appreciate you to give feedback.
You can also leave comments under this thread if you want to give more specific things

My understanding is than slack will delete everything older than one year not 90 days and this only for the free version. https://slack.com/help/articles/29414264463635-Updates-to-message-and-file-history-on-free-workspaces

*Thread Reply:* Sorry yes, 1 year. But we are on the free version so would still affect us
*Thread Reply:* Are you sure? Because i can see 2023 content in this channel..
*Thread Reply:* We are currently on a 1 month trial 😅
*Thread Reply:* So not surprising 😬
*Thread Reply:* We are definitely not paying for anything
*Thread Reply:* Oh i see 😁
If moving to element is going to happen then consider exporting the workspace info could be useful https://slack.com/help/articles/204897248-Guide-to-Slack-import-and-export-tools

*Thread Reply:* Yes, I already do this every time we have a pro trial 🤣
@channel

 }
}

 }
}

 }
}


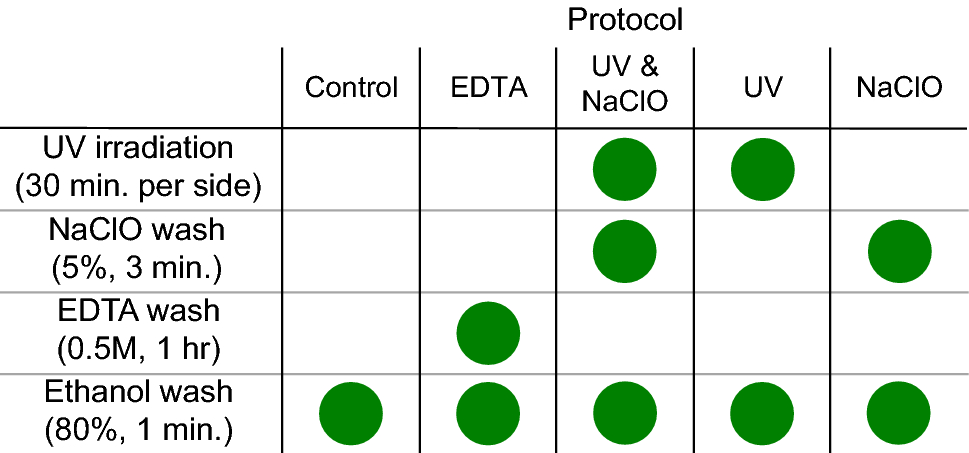

 }
}




 }
}
 }
}

 }
}