1 Double stranded DNA libraries


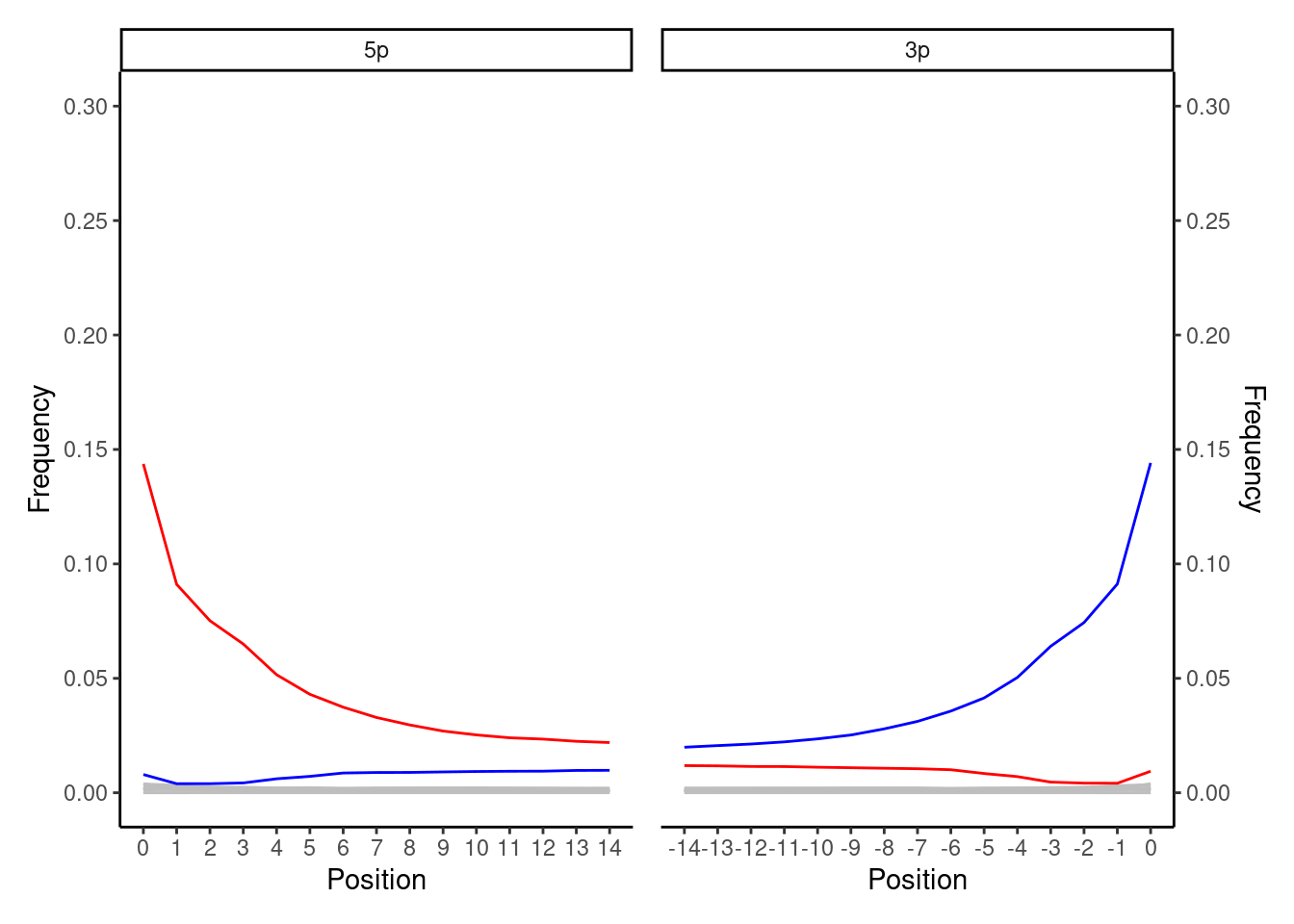
This is the ‘classical’ ancient DNA plot that you will see most often in palaeogenomics. You expect to see a smooth curve from the beginning of the read (position 1) to a flat line in the middle (e.g. positions 10-25 in mapDamage plots). At the 5’ end this will be indicated by the original C to T deamination, whereas the 3’ of the molecule will show the complementary G to A. You only see deamination the C to T (and complement G to A) at one end of the the molecule, as during typical double-stranded library construction protcols (Meyer and Kircher 2010) only one end of the single-ended overhangs of a DNA molecule is repaired by being ‘filled in’ (where the mis-reading of the deaminated C occurs). Overhangs at the other end of the molecule (which may also hold cytosine demination) are ‘blunt-ended’ by being trimmed off. Both fill-in and blunt-ending reactions are performed to allow ligation of next-generation-sequencing adapters and/or internal barcodes to both ends of the molecules. The highest frequency point of the curve can vary from 1% to 50% depending on the age and preservation of the sample.
If you get such a plot with smooth lines from ancient DNA double-stranded libraries, this is a good indication you have authentic ancient DNA!