Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
/github subscribe SPAAM-workshop/AncientMetagenomeDir
@Sterling Wright has joined the channel
@Antonio Fernandez-Guerra has joined the channel
The general layout as I envision it currently can be seen here: https://github.com/SPAAM-workshop/AncientMetagenomeDir
A lot of it is still up for discussion, so we can tweak based on your feedback and so on.
I've tried to write a lot of documentation, so please look through it and see if it makes sense, and give it ago. I've already made issues for papers that need to be added if you have nothing off the top of your head (but this is only from my knowledge/context curretly, so feel free to add more).
@James Fellows Yates I will take the standards.tsv and make it mixs compliant, then integrate the new fields a extensions for MInAS
*Thread Reply:* Ok, please make a PR so I can check! I want to be careful that it is just for consistency within this database.
I don't want it to get very large and inflexible, as that is not the purpose of this!
*Thread Reply:* But of course making it as portable as possible would be awesome
*Thread Reply:* The ideal would be to start adapting the standard that is going to be part of GSC and hopefully adopted by INSDC
*Thread Reply:* OK. I'd like to have a look. Maybe most columns are fine with that but then some could be potentially problematic (cultural era, dating etc...), 'cause Archaeologists 😉
*Thread Reply:* Have you already got tihs defined anywhere public?
*Thread Reply:* The repo is awesome, I will try to talk this week with Guy Cochrane (head of ENA) if we can start a mini project to integrate this
*Thread Reply:* Is not open yet, but the core descriptors of mixs are open
*Thread Reply:* Once we are also a bit furhter along, I'll try to add some CI checks, so we can check PRs against the standards.csv file(s)
*Thread Reply:* Those are the ones you should first encourage to comply
*Thread Reply:* Those have to be obligatory in the tsv file
*Thread Reply:* Ok. Yeah, then please make a PR with those proposals and definitions
*Thread Reply:* I will invite you to the doc I am working and then I can do the PR
*Thread Reply:* If doc is google drive: jfy133@gmail.com
Please also give me your github handles and I will add you as members
(to the whole organisation that is!)
@James Fellows Yates awesome job!
@Ele if you have time at 14 CEST (I guess 13:00 in York) this Friday, I'll be doing a intro to git with my supervisor (we will use this as an example). Let me know if you would like to join
*Thread Reply:* Sounds brilliant I would love to join - thanks very much!
And ping @Shreya. I can't remember exactly where you are in the US, but Tina will be joining from the US too so might not be too awful
*Thread Reply:* Hi James, thanks for including me— excited to be part of the project! I’m very much a beginner with git and would love to join in on the intro. I’m based in Chicago so 14CEST works for me!
/github unsubscribe spaam-workshop/AncientMetagenomeDir commits
@Ele @Shreya @Becky Cribdon only thing I would like you to do before Friday is set up a GitHub account and send me your username, so I can add you to the SPAAM organisation. Everything else we will go through
*Thread Reply:* thanks James-- I’m shreyaramachandran
*Thread Reply:* Thanks, I'll be there as Allaby-lab.
*Thread Reply:* Is your name associated with the account? As in the display name?
*Thread Reply:* Just asking as that would be important for you getting correct attribution
*Thread Reply:* Thanks James - I have set up an account under eg715. Looking forward to tomorrow
*Thread Reply:* You should've recieved an invitation to join the organisation!
@Ophélie Lebrasseur has joined the channel
🎉 thanks to @ivelsko for the first submission!
Review requested for: https://github.com/SPAAM-workshop/AncientMetagenomeDir/pull/21
@Anneke ter Schure has joined the channel
@Anneke ter Schure @Abby Gancz @Jonas Niemann Very sorry I saw your responses in #general so late. I will be running the very basic intro to GitHub at 14:00 CEST *TODAY*. I will post the link here
Also @Jonas Niemann congrats to you and Theis, your birch pitch paper is the *First* that I've found has uploaded their data to ENA with correct metadata 😉
@Anneke ter Schure @Abby Gancz @Jonas Niemann if you can make it today, please send me your github usernames before then, please!
@channel for this who want to join the GitHub tutorial at 14:00 today, this is the video channel (chrome/firefox preferred!): https://meet.gwdg.de/b/jam-m3j-kyj
If you've not already, please send me your github username (so I can add you to the organisation)
@channel If you have problems joining please ping me here!
@Becky Cribdon we are starting
https://meet.jit.si/SensibleSpendingsFinanceThere
I'm sorry guys, I got the time difference completely wrong!
*Thread Reply:* Hey Becky, no problem. If you can wait 20 minutes I can go through with oyu privately if you want
*Thread Reply:* Or just message me here if you look through the slides
*Thread Reply:* Yes, I'll have a look and let you know if I have any questions. I hope it went well. Oops!
*Thread Reply:* Surprisingly well!
*Thread Reply:* Whcih is a good sign I hope 😆
*Thread Reply:* Okay, I added Fagernas 2020 and requested your review, but now I'm getting emails saying it has failed tests. Let's discuss when you have time.
*Thread Reply:* Will do! Tests are new, might need tweaking
*Thread Reply:* I just had a look
*Thread Reply:* Try taking out the umlaut on the a
*Thread Reply:* Might be a limitation of the Regex atm
*Thread Reply:* Good shout. I think it's gone through now?
*Thread Reply:* It did indeed!!! :mask_parrot:
*Thread Reply:* I'll do the manual check later!
*Thread Reply:* Either tonight or tomorrow, toddler allowing
@Christina Warinner has joined the channel
For those who might be already working on PRs: we now have working validation checks when a new PR comes in. So if you get a ❌ you'll need to check the error by pressing the 'details'. The error it self might have a lot of weird stuff but in the middle of it you should be able to see the error.
For example in here: Fagernäs2019, is not allowed as it has an invalid character (ä). In his case, make a new commit to just replace it with the character without the accents. This is unfortunately a limitation of Regex (due to a very complicated problem, so this is a compromise)
Thank you to @Maxime Borry for making the extensive case against the special characters ;)
/github unsubscribe SPAAM-workshop/AncientMetagenomeDir deployments
@channel anyone willing to do reviews for the following for me: https://github.com/SPAAM-workshop/AncientMetagenomeDir/pull/24 (7 samples)
https://github.com/SPAAM-workshop/AncientMetagenomeDir/pull/21 (~40 but most are from the same site)
Basically just need you to check the metadata info looks approximately right (ignore the minor changes)
@Vilma Perez @Abby Gancz @Jonas Niemann @Ele @Shreya I'm adding a lot of papers for you guys to try out of you want 😉
/github unsubscribe spaam-workshop/AncientMetagenomeDir
@channel I've unsubscribed from the github stuff. Sorry for the constant pinging, was still exprimenting!
@channel Just did a vomit of all pathogen papers I could think of/find: https://github.com/SPAAM-workshop/AncientMetagenomeDir/issues
We now have 60 open PRs, so feel free to start assigning yourself!
@Maria Spyrou wrong channel
Sorry @Miriam Bravo I forgot to add you earlier as :face_palm: my bad
*Thread Reply:* Thanks James!! 😄 . Sorry for not answering earlier.
*Thread Reply:* No problem. Feel free to ask any questions if you want to catch up
*Thread Reply:* I will reach out to you as soon I finished reading all the comments . Thanks!!
Hi everyone! I have heard about this project and though a study we recently published about historic herbarium metagenomics (samples were collected up to 150 years ago) might be interesting for it although the samples are not ancient. What do you think?
*Thread Reply:* For clarification, I consider anything ancient being from anything not currently living ;)
Would you consider the data to be microbiome?
Or should we make a new table?
Hmm, it is microbiome, but I think we should make (a) new table(s). To broadly cover the sample types I can think of, something like ‘microbiome-animal’ and ‘microbiome-plant’, and perhaps changing ‘sediment’ to ‘environmental’ and adding ‘food’ for things like scale in pots
Thanks for bringing this to our attention @Vanessa Bieker 😊 I agree with @ivelsko that it would be good to add one or several more tables to encompass the diversity of ancient metagenomic samples.
@ivelsko so we should rename the current ancient-microbiome to be more specific? Should we then do the same for pathogens? I'm leaning towards agreeing but don't want to result in ever smaller fragmentation...
*Thread Reply:* Yes, I think that’s the way to go. But I agree we need to limit the number of categories. I think the 4 categories I mentioned cover everything I can think of (for example parchment could go into either microbiome-host or environmental)
Would it make sense to have one for human + animals and one for plants+food scale+other?
We could do what I did for pathogen: domain
Or family or whatever in this case
(if it's still falling in microbiome)
I wouldn’t break down pathogen too much, b/c that’s a much tighter focus
I would use host-associated or not and then environmental to all what you decide not to be in the your groups of interest
Using the controlled vocabulary from environmental ontologies should be a good compromise
If not we will be inventing new terms that already exist
@Antonio Fernandez-Guerra fair point. I will explore to see if there is any useful categories.
Host associated is not so much the issue here though, I think it's more plant people Vs mammal people (so to say)
So it's about accessibility
Archaeologists Vs ecologists, for example
This were the material takes place
And don’t forget microbiology
At the end of the day we are doing microbiology, and this has very specific concepts we should try to follow
*Thread Reply:* Oooh careful there. I wouldn't consider myself a microbiologist ;)
*Thread Reply:* A lot of ancient eDNA focuses on fauna and flora, not microbes
*Thread Reply:* But most of the papers in the PRs are microbiome related
*Thread Reply:* Also environmental ontologies cover non living entities
*Thread Reply:* Only because they are the people in pestering distance from me ;)
*Thread Reply:* But yes, ok
I wouldn’t consider food microbes/microbiomes to be environmental, since they’re very specifically different from environmental samples, even if many of the communities are inoculated from the environment. @James Fellows Yates when you’re looking at the environmental ontologies do they have some way of distinguishing food vs environment? That’s a distinction that will be important to anthropologists/archaeologists. It’s not intuitive to lump food scale in pots with ice cores even though they’re both not host-associated
@ivelsko yes, you can be very specific
I would select few representative terms that suits the project
Then you have already put data in a reasoning system
Then for example from EnvO you can go to FoodOn (https://www.nature.com/articles/s41538-018-0032-6)
I believe that if we can integrate what is being doing here in the larger picture we are shaping a FAIRer future for our data
Also we can ask for new terms if needed, the main developer of EnvO is a good friend and he told me that he is happy to help
Ok. I think we are getting there. I'm starting to see a possible strategy (but I'll check against the onyology of course)
- host associated (microbiome-like)
- clinical (pathogens, this us a special case because only really referring to single genomes)
- environmental (sediment, etc)
- manufacturing/industrial (food crusts, parchment, artefacts etc)
How does that sound @channel
And then I guess @Vanessa Bieker's herbarium samples stays with host associated.
Only problem maybe is if people do decompositional studies of skeletal elements, but I guess that would still fall under environmental. But we need a very clear way of separating that from the sediment stuff.
I will have a look on Monday to the envO terms and provide a list we can choose
We can also give a try to extract https://extract.jensenlab.org and see what we get
@ivelsko I've reviewed Brealey for you, a few minor structure/consistency changes but the metadata looks all correct to me
(please request re-review whne I know to look again!)
*Thread Reply:* I updated the file just now, can you review it again?
*Thread Reply:* Will do! Just tweeting aobuti t 😉
*Thread Reply:* (the project not the PR)
*Thread Reply:* Just nee dto fix something. NAs don't work for strings in JSONs unfortunately 😞
*Thread Reply:* I'm using 'Unknown'
*Thread Reply:* I’ll put that in instead then and make a new commit
*Thread Reply:* That's only for strings
*Thread Reply:* LatLon is fine with NA I think
*Thread Reply:* Updated with Unknown for site and geo_loc
*Thread Reply:* Last thing:
Gb1-reg should be Gb1 now. And then we can merge 🎉
If anyone wants to spread the word: https://twitter.com/jafellowsyates/status/1292758485134868482
@Alex Hübner PR review ready for you too
So I've been hunting through a lot of the ontologies listed on the OSL, but none of them come really close unfortunately 😕. A lot of them are highly specific to specific sub-disciplines or seemingly abandoned.
However, I would like to propose the following based on an approximate mish-mash of a few of them. From there were can start refining over time (or even make our own 😆)
ancient-microbiome: host-associated multi-community (calculus, paleofaeces) ancient-pathogen: host-associated single-population (pathogens genomes, if in a community sample can also be listed in the ancient-microbiome list) ancient-environment - natural environments (sediment, soil, icecores) ancient-anthropogenic - human (highly)-modified/created environments (pot crusts, artefacts, parchments, middens, latrines)
One material type I'm unsure about is burial/grave sediment/soil. This has been argued to still contain traces of the human microbiome. But it's mostly soil so maybe nevironment fits, but it's not 'natural' if we consider it may have been in a grave with human artefacts (metal, wooden coffin etc). Another one I'm unsure about is metagneomes derived from seketal material (e.g. from a jaw bone). This will mostly reflect the burial environment but it is skewed to microbial taxa that can colonise bone...
The names are used for readability, but the descriptions follow common ontology categories. But also, importantly for accessibility, this covers I think the main ancient metagenomic 'sub-fields'.
@channel what do you think? Do you feel you can slot your personal research area into one of the four?
*Thread Reply:* > One material type I’m unsure about is burial/grave sediment/soil. This has been argued to still contain traces of the human microbiome. But it’s mostly soil so maybe nevironment fits, but it’s not ‘natural’ if we consider it may have been in a grave with human artefacts (metal, wooden coffin etc). Another one I’m unsure about is metagneomes derived from seketal material (e.g. from a jaw bone). This will mostly reflect the burial environment but it is skewed to microbial taxa that can colonise bone... For this we should ask Pier, he will give us a good advice
*Thread Reply:* Can you do that?
*Thread Reply:* yes
I think ancient microbiome should have another name (I don’t know which) We are working with ancient microbiomes of very different origins like deep sea, permafrost, carbon reservoirs, human/animal related, and all are microbiomes and ancient
*Thread Reply:* That's very embarrassing, I've been complaining that loads of gut microbiome papers don't specify that in the title, they just say human micrbobiome. So I get excited and rapidly disappointed when it's all poop.
But clearly I've been sucked in 😢
*Thread Reply:* Nothing to be embarrassed, most probably many that use microbiome in the title they refer to the microbiota...
I don’t know, maybe ancient host-associated microbiome let’s see if there are other suggestions
*Thread Reply:* Maybe have to, even if it's ugly
*Thread Reply:* When you are looking at deep seq/permafrost, it's definitely just(/primarily) microbes you're looking at, right?
*Thread Reply:* The whole community
*Thread Reply:* Wondering if we could separate host-microbiome vs envionrmental-metagenome
*Thread Reply:* But also eukayrotic free-DNA?
*Thread Reply:* You have a mix of the different dna pools
*Thread Reply:* Yeah, which to me would be metagenome rather than just microbiome
*Thread Reply:* But of course same applies to 'host-associated' microbiome
*Thread Reply:* I would have a category as environmental-microbiome or similar as there will be people focused on this, i.e. looking for spore-related studies or recovering frozen microbes/viruses
*Thread Reply:* I would use the definition of microbiome from here: https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-020-00875-0

*Thread Reply:* I wonder if then we are getting to specific then. Given shotgun contains not just microbes....
maybe ancient-hostassociated-metagenome ancient-hostassociated-pathogen ancient-naturalenvironment-metagenome ancient-anthropogeneic-metagenome
or something
*Thread Reply:* Yep, I think this would be the most general
*Thread Reply:* then we cover all cases and we are not limited to being microbiome
New suggesiton after talking to Antonio
- ancient-hostassociated-metagenome
- ancient-pathogen
- ancient-metagenome-naturalenvironment
- ancient-metagenome-anthropogeneic
(terms maybe not in that order)
I just realized that using the term pathogen isn’t quite a big enough umbrella. For samples that are capture of a species that isn’t necessarily a pathogen, or isn’t acting pathogenically, like what I’m working on, would that go under pathogen even though it’s not a pathogen?
Yes it would technically. It's basically single genome
But we want to specify it's microbial I guess in that case...
maybe we can get something from here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6786532/
I will ask Luke about the parasite-associate package
- ancient-metagenome-hostassociated (essentially original host microbiomes from skeletal or mummified material)
- ancient-singlemicrobe-hostassociated (microbial genomes, primarily pathogens)
- ancient-metagenome-naturalenvironment (soil, sediment, non-microbiome bones)
- ancient-metagenome-anthropogenic (parchment, artefacts, non-host herbarium, storage buildings etc.)
As part of the sediment crew, I like the look of this.
Burial soil is a tricky one, but I would lean towards anthropogenic myself. Other human-associated samples will have a lot of environmental input too.
*Thread Reply:* Sediment crew 🤣.I like it. If we ever get money to pay for slack I'll make you a team with that name
@channel the lists have now been restructed as above!
anthropogenic will need modification I think but we wait until someone submits papers for that
(@Alex Hübner this restructure mainly affects your current PR, but I've made a PR into your fork which should update everything - but I didn't make the changes as noted by my review)
@channel now we've got some data, anyone want to to play around with making a few summary graphs to go on the website: https://spaam-workshop.github.io/AncientMetagenomeDir/
I just started look at Mühlemann 2018 Hep B. virus paper - they only published their 'assemblies' , not the raw sequencing reads - my question for the pathogen peeps: should I include a column saying whehter raw sequencing data is avaliable?
For today at 14:00 CEST: https://meet.jit.si/AncientMetagenomeDir
Slides: https://hackmd.io/@jfy133/B156Z2mfP
I've changed tweaked the definitions of the list, as I was coming up with more problems.
hostassociated metagenome list basically includes any tissue or original contents derived from a body (skeleton or mummy). This therefore can include teeth or bones etc.
the environmental is therefore 'sediment/soil' like samples (which is easier to define)
It sounds good to me, we also need to think about the issue @ivelsko raised about pathogens (https://spaam-community.slack.com/archives/C0183TC8B0R/p1597061759074900) maybe something like parasite-associated or similar. I don’t know how much this can be useful https://github.com/GenomicsStandardsConsortium/mixs/issues/65 or https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008028 for this discussion
That definition is now (for the time being) just any microbial genome, commensals are fine. Basically anything old and has been reconstructed to a genome level. It was originally called pathogen because 95% of ancient microbial genomes have been those. In the future we can review the species namesto assign extra tags to indicate pathogenicity
Then I would remove the commensal part in the description on GitHub, this is just one of the many potential ecological interactions one can get for a single genome
I've kept it in for 'accesisbility' of aDNA people who don't have microbiology backgrounds, but put an improved clarifying statement with (eg. not just pathogens but also commensals)
👍 now reads better
@channel OK new problem. The Pratas 2018 paper where they do metagenomic analysis of an ancient polar bear jawbone isn't actually with fresh data, but reusing previously published data (used for WGS, and didn't do metagenomic analysis). My question is how to deal with this:
1) exclude this publication (as if we include samples from WGS then we would have to delve into the hundreds of ancient human pop gen studies)
2) include it (with new definition of papers to include in the 'Dir being had to have some form of metagenomic analysis), and include another column called original_data_doi or somthing?
If we do include we definitely need something like 2), to ensure correct attribution. But also I don't want to see column creep...
Thoughts?
I agree that we don’t want to include all the (human) popgen papers that have uploaded raw data that could be used for metagenomic analyses. Adding something like this where metagenomic analysis was actually done is fine I think
I agree with Ivelsko.
Also, do I remember a README somewhere saying that re-analyses of previously published data were fine? Presumably that was for when both the new and old analyses were metagenomic, but could we use the same system for Pratas 2018?
That was newly re-sequenced data of a previous sample
i.e. Pratas did no lab-work at all
With the assumption is that the originally sequenced data would already be in our list under a previous paper, e.g. like Ziesemer 2015 (that @Ele is preparing) -> Mann 2018
But ok, then maybe:
original_data_doi - DOI of original publication, where the sequencing data of the current study performing metagenomic analysis was generated for other purposes (e.g. host genome analysis) in a separate publication
I would be pragmatic and follow what is in the README:
AncientMetagenomeDir is a community curated resource of lists of all published shotgun-sequenced ancient metagenome samples.
if Pratas 2018 did not submit any data to ENA/SRA I would not include it to the list. Then things can get complicated with different exceptions we might find.
> if Pratas 2018 did not submit any data to ENA/SRA I would not include it to the list. Ooh that's a nice definition
You might have different levels of DOIs before you get to the original_data_doi
better keep it KISS 🙂
As @ivelsko says, as it would reduce a risk of someone requesting the addition of re-analysis stuff I guess...
Only caveat is many of the singlegenome papers already are using ENA/SRA data from human pop-gen stuff (even if published in a separate publication).
But I guess that list will always be a special acception
Ok, cool. I'll then close that and maybe clarify in the READE
For the single genome I don't think it is a big issue since most of the times it requires resequencing of the data or additional capture and human pop-gen studies only tend to put the human mappint reads in the ENA/SRA...
Another pathogen/single-micorobe-genome question: To what level does a genome have to be ‘complete’ in order for it to be included on the list? Papers often report some complete - or almost complete genomes, as well as samples that are maybe only 50% covered at 1-fold, as well as samples that can be positive based on a few thousand SG or capture reads, but it’s not enough for in-depth analyses.
Maybe we could have another column indicating completeness?
Aida asked me the same thing. I sorted of said enough SNP data to perform some form of whole-gneome phylogenetic analysis
Completeness becomes subjective though 😕 (or something people can argue about, so to say)
Have you across a problematic sample?
Yes, but some people do phylogenetic analyses on genomes with less than 50% coverage at 1-fold, while others would choose not to do that
Ok fair enough. I think I say >50%?
*Thread Reply:* Majander 2020 has a genome of 46.89% covered at 1-fold
*Thread Reply:* OK, then 50% is a bad idea
*Thread Reply:* That indeed is arguably enough
*Thread Reply:* It also depends if you have the correct ref genome to begin with etc.
*Thread Reply:* Again, up to the re-analyser to evaluate ;0
no, you said whatever it get reported as a genome in the paper...
I would honestly take the same approach. It's the same as we agreed on for the microbiome samples; if it was published as one we make the 'assumption' peer-review should've filtered out crap. Of course it doesn't always, but this list is not meant to tel lyou waht is good or bad
Just what you could try to reanalyse
Maria Spyrou and I decided that we will consider something a positive/evidence of plague when it has 0.1X coverage
also it depends what people want to do with the data
if it is for analysis you may just want to use good quality data
if you just wanna plot where potential positives have been found, then I'll argue we put in whatever people have reported
and since it is just supose to be a list of resources, people can then read the paper and determine what data they use?
OK yeah, as @Antonio Fernandez-Guerra sort of argued earlier - lets keep it practical. If it's reported in a paper as a 'genome' you can include it. The important thing is that it's on the chromosome not just single plsamids (like Schuenenmann PNAS... 2011, I think)
I would add the coverage information more than if complete or partial, this can be a long discussion, and especially when you plan to add MAGs
But people don't report coverage consistently... that would be a nightmare
this is what we are working for
to get a set of best practices
for example this is well charactized in MIGS https://www.nature.com/articles/nbt1360
As this has started arguing amongst ourselves alreayd, I think we should go to the fall back of 'if it's reported as...'
I would follow the same principle of the Pratas 2018, if it is a field that needs a lot of thinking is going to be a nightmare to mantain
OK, so practical example: In my 2018 Salmonella paper I have 5 samples that I only did basic SNP analyses for to confirm that they were positive, some are as high as 1.2X and some as low as 0.36X, should i include all of them then? even though I only really fully analysed the 5 high coverage genomes?
Should be inclusive until we collective agree on standards
in the case you still you want to report this, it should include the reference_accession and maybe mapping_method and then get a table like this https://www.nature.com/articles/nbt.3893/tables/1 with the thresholds you agree
in MInAS we will cover all of this
Using this table as example, we can brainstorm if only coverage is a good way of saying if something is positive or not, specially when the average coverage a genome recruits can be so low
then this can be used as a guideline in the future
Hey all, I’m getting a bit confused about recording the latitude and longitude of the sites. Are you specifically looking up the site location? For example one sample I am looking up now is from Middenbeemster. I have looked up the reference to the site, it is https://www.sciencedirect.com/science/article/pii/S1879981713000831 but I don’t see any more information here about the specific site location. What’s the best thing to do?
*Thread Reply:* Try get it as close as possible
*Thread Reply:* Just whilst you’re here James, I am only recording shotgun sequenced data right?
*Thread Reply:* I normally go in the following order:
1) if there is lat/lon given, use that 2) if there is a map, approximate on Google maps 3) if there's is a local town or something, or the site is from a site where there, just put the town name 4) if it's not really clear, put it in the middle of the region but only give 2 decimals
*Thread Reply:* Correct, only shotgun
*Thread Reply:* For middenbeemster you can use the same values as Mann 2018
*Thread Reply:* That is super helpful - thanks! Mind if I copy that to the notes I am writing up?
*Thread Reply:* They are the same samples
*Thread Reply:* Feel free to copy everything I say
*Thread Reply:* Certainly better than my own memory 🤣
*Thread Reply:* Brill thanks! Hahaha, same I have to write EVERYTHING down
*Thread Reply:* Like I said if you could it on the wiki that would be awesome (not obligation though)
*Thread Reply:* Of course, happy to share 🙂 I’m making my way through one then it would be helpful for someone else to cast their eyes over it too
*Thread Reply:* I just merged the current master into your Ziesmer-2015 branch so you don't get too far behind
*Thread Reply:* I just saw this - thanks!
I've started a wiki page to summarise some of the discussions/conclusions from slack: https://github.com/SPAAM-workshop/AncientMetagenomeDir/wiki/Discussion-Notes-on-Definitions
Feel free to edit/extend as neccessary
@Ele I would recommend making a new page for your notes, we can discuss a title when it's ready
*Thread Reply:* Great - thanks. Hoping I’ll be done pretty soon will let you know
@channel there are still a lot of pathogen papers open, so feel free to join in 😉
Ok, I've found 2 single-genome papers now that have just published consensus sequences (yay reproduible science >.>) and not raw metagenomic data. We've talked briefly in the past and for this context we shouldn't exclude these because it falsely makes it seem there are less ancient microbila genomes than there are.
I propose a new column for the single-genome list only called: datatype, which can have two values: raw_data and consensus_only. The archiveaccession can then be the genbank accession ID of the sequence. What do you think. In particular @aidanva @Åshild (Ash) @Maria Spyrou @Shreya @Miriam Bravo who I know work on pathogens
*Thread Reply:* I agree, it would be useful to have column listing rawdata/consensusonly. If both are provided, do we list just raw_data? it might be useful to know if both are available @aidanva @James Fellows Yates
*Thread Reply:* Suggestions on concise way to code this?
*Thread Reply:* unless we compress
*Thread Reply:* so: raw, consensus, contigs, mag
*Thread Reply:* Ah but then that's not following the suggestions from Antonio 🤔
*Thread Reply:* but the suggestions from Antonio were dealing with organelles, plasmids and others right?
*Thread Reply:* Oh ffs :face_palm: sorry I'm all mudlded today
*Thread Reply:* correct yes
*Thread Reply:* OK back at it
*Thread Reply:* raw, concensus, contigs, mags
*Thread Reply:* And then combine them as needed
*Thread Reply:* but I think what ash is referring is how we report then the data, so shall we report consensus + raw data for example in the archive_accession
*Thread Reply:* rawconcensuscontigs
*Thread Reply:* But archive_accession should still be at sample level
*Thread Reply:* And that stuff should all be linked at the sample level
*Thread Reply:* it may get messy if people uploaded raw data in one place and contigs in another?
*Thread Reply:* True but that's stil up to the researcher to find
*Thread Reply:* I am just thinking then one will have to mention more than one archive as well
*Thread Reply:* > True but that's stil up to the researcher to find
*Thread Reply:* sure, but wouldn’t that mess up the checks that you do to see if the data complies to the rules?
*Thread Reply:* I don't think so
*Thread Reply:* The archive_accession is currently free text
*Thread Reply:* just checking the code, and it shouldn’t
*Thread Reply:* ok, I think adding the additional column makes sense, and then we agree to have a column with data_type or similar name, with potential fields being: raw, consensus, contigs, mag
*Thread Reply:* how will you define the difference between contigs and mag?
*Thread Reply:* Yeah realised that's the same thing
*Thread Reply:* Or we could just call it assmebly?
*Thread Reply:* I think we always have mags really…
*Thread Reply:* yeah, I think assembly is better
*Thread Reply:* And hierarchy: raw < assembly < consensus?
*Thread Reply:* I don’t understand the hierarchy…
*Thread Reply:* assembly and consensus are often used interchangeably in the literature
*Thread Reply:* How does one seaprate a colletion of contigs (an assembly to me) vs a final fasta
*Thread Reply:* if the consensus is called from an assembly, it is sometimes referred to as an assembly.
*Thread Reply:* we could provide definitions of what each means in the context of our db
*Thread Reply:* I woul dopt for that
*Thread Reply:* But maybe you guys can agree on the least ambiguous terms?
*Thread Reply:* Sure, @aidanva shall we set up a singlemicrobe channel for us and others to discuss this stuff?
*Thread Reply:* Sure, let’s be the god of the pathogens, and decide on the terms. Do you wanna set up the channel?
*Thread Reply:* Maybe founders of rather than gods 😉
*Thread Reply:* Please also announce it on #general once made!
*Thread Reply:* gods
I think it makes sense to do so
Are there any other categories you can think of?
Or types of data that might be produced?
I mean… maybe people will also be interested in knowing if the genome was reconstructed based on shotgut or capture data?
*Thread Reply:* This is the thread @James Fellows Yates. Maybe this column is not necessary, but in terms of mixture we could just say “shotgun/capture” or something like that
*Thread Reply:* Yeah. Ok, but I think my original point still stands
*Thread Reply:* Was thinking ahead, my ba
*Thread Reply:* Let’s no add it now, but maybe something to consider
That's library level though. I mean in terms of data types
and I am still trying to wrap my head around what to do with plasmids, or organelles, because some of them may be revelevant for some questions like: evidence of X genome in the past
(and I want to avoid that mess for the time being 😅 )
For that maybe you could make a 'pathogen' committee to discuss that. I am familiar enough with data that I can help decide that, but stuff like plasmids/organelles I dunno much
Feel free to make a new channel
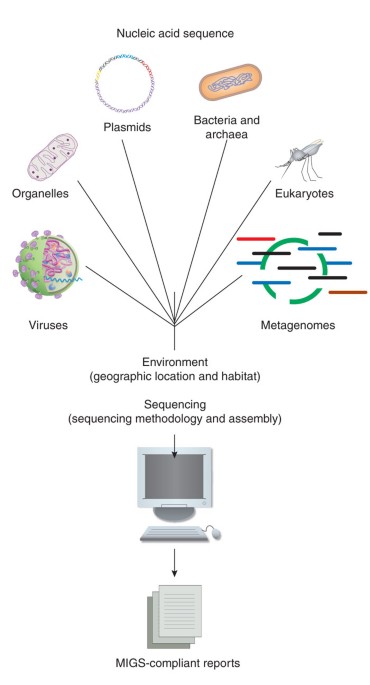
for organelles, plasmids and others I would stick with the investigation_type from MIxS: https://microbiomedata.github.io/nmdc-metadata/docs/investigation_type.html
*Thread Reply:* Looks very nice, I think that would be easily incorporated as a column
*Thread Reply:* OK. @aidanva you can get a 'consensus' on what is allowed to be included, we can add this as well
*Thread Reply:* Although we need to work out how to define if it is a mixture...
*Thread Reply:* if a sample has shotgun and capture data
*Thread Reply:* Remebmer we are listing samples
*Thread Reply:* (as far as possible, of course)
*Thread Reply:* yes, many of the things that can be related to a modern microbiome, most probably is already included in MIxS/MIGS/others
*Thread Reply:* I would use the terms of MIxS and coin our specifics
*Thread Reply:* Sorry ignore me about the mixutre thing
*Thread Reply:* Was on the wrong thread
*Thread Reply:* thinking on wrong thread**
*Thread Reply:* (the list Antonio sent has stuff like: single amplified genome, single-cell, metagenome-assembled etc)
*Thread Reply:* we may have to coin specific terms for our list
*Thread Reply:* but there are terms that we can definitely reuse
*Thread Reply:* I always would try to use one of the existing terms, and if not discuss potential new ones
*Thread Reply:* Yes, just need to be careful we are not using it with different meanings that may mislead people
*Thread Reply:* we don’t want to overload the list
@channel We have now created the channel <#C0193SH0KEF|ancient-single-genome> where we will discuss things specific to the ‘ancientsinglegenome-hostassociated’ list. That way fewer people get spaamed with niche discussions 😉
Hi all, I am correct in thinking that if I have a paper which look two samples from the same individual and did multiple sequence runs, this counts as 2 sample entries (despite being from the sample ind) and I put the run accessions in the archive_accession column separated by a comma?
*Thread Reply:* Mm, you shouldn't need run accessions
*Thread Reply:* YOu need the sample accessions
*Thread Reply:* If they are two separate samples (even if from same individual), they can be two lines
*Thread Reply:* Ok great thanks, yeah there are only 2 different sample accessions, but multiple run accessions — just go with sample accession?
*Thread Reply:* Yes, we only ever use sample ones because run accesisons get messy (that'll be project nr 2 ;))
*Thread Reply:* Brilliant - thanks for the help 🙂
Hey, would someone please mind adding Borrelia recurrentis to the enumns/singlegenome_species list? (or can I do this?) Thanks 🙂
Thank you, that would be great 😄 I wasn’t sure if I was allowed 😂
I think as soon as you follow the NCBI Taxonomy, every one is allowed to add new species, and if you made a mistake, someone will catch it during review 😉
ping me, I’ve got a bit of time this afternoon to do it 🙂
*Thread Reply:* Hey sorry for all the questions, I am still failing the checks due to it not recognising Borrelia recurrentis. In the list the error is giving Borrelia recurrentis is not listed, but it is present in the enumns/singlegenome_species list (thanks for adding it 🙂 ) is it just taking a little time to update? Do you think it’s best to wait for a while or should I remove the PR and make a new one?
*Thread Reply:* yeah, I realised I should do a pull request on main, because that it is where it checks
*Thread Reply:* I asked James to review it for me, so hopefully when it is merged your test can pass and then we can merge your PR
*Thread Reply:* the checks will run automatically once it detects a change
*Thread Reply:* Oh great, in that case sorry I made new commits that weren’t really needed
*Thread Reply:* no worries, I am also figuring this git stuff
*Thread Reply:* btw, I left some comments of changes you should do to the table
*Thread Reply:* not sure if you have seen them
*Thread Reply:* Thanks for the comments Aida I have made the changes 🙂 Would you mind explaining the differences in the accession codes please? I was using this link: https://www.ebi.ac.uk/ena/browser/view/SAMEA4771255 I can’t see the ERS2591211 code anywhere here, should I have been looking somewhere else?
*Thread Reply:* @Ele yeah it's annoyingly hidden
*Thread Reply:* I also couldn’t find it at first, but when you are in the project page, you can open the show columns section bar, and you need to tick ‘secondarysampleaccession’
*Thread Reply:* Top right press 'read files'
*Thread Reply:* Then open the white show column sleection in the middle of the page,
*Thread Reply:* First column, 2/3rds of way down, tick 'saecondarysampleaccession`
*Thread Reply:* Argh! Yep the secondary thing. Got it sorry - this is all quite new for me. I thought I had checked that - maybe I reloaded page!
*Thread Reply:* No worries! I added the new species to the list, and reexecuted the tests so you should see a nice ✔️
*Thread Reply:* Why are we using the secondarysampleaccession @James Fellows Yates?
*Thread Reply:* It's more closely linked to the data itself
*Thread Reply:* The SAMEA-like codes are a part of this BioSample database which I think allows linking to other types of data (transcriptomics, proteomics etc)
*Thread Reply:* @Ele you can merge the pull request now!
*Thread Reply:* But in the the end goal would be to make tools that allow you to automatically download all the sequencing data you request
*Thread Reply:* (all non-UDG treated data from Finland from 1300-1700 year ago)
*Thread Reply:* Or even have automated taxonomic profiles etc which you can download for comparative data
*Thread Reply:* @Ele merge merge! It's an awesome feeling and then to delete the branch 😄
*Thread Reply:* you can also merge @James Fellows Yates
*Thread Reply:* Ele jsut did it
*Thread Reply:* @Ele if you didn't link the PR, please also clsoe the issue (if you've not already done it)
*Thread Reply:* I just clicked and the post arrived which delayed my celebration messages!
*Thread Reply:* Sorry James not really sure what you mean about the linking. I have a prompt to delete the branch… Should I do it?
*Thread Reply:* Linking is separate! But yes, you can delete the branch
*Thread Reply:* As you've added everything from the paper
*Thread Reply:* For the linking:
*Thread Reply:* Wow that feels very satisfying. Thanks both for talking me through 😁
*Thread Reply:* An example from another PR
*Thread Reply:* If you start # (if not at the begining of hte line), it'll give you list of issues and PRs and if you select one, will be rendered as a hyperlink. When you open the PR someone an then quickly find the issue to check for other issues.
A nice trick with github, is if you put certain keywrods like 'close', Github will automagically close the issue for you once the PR is merged
*Thread Reply:* Also once you link it, the issue will be updated and the PR displayed on the issue
*Thread Reply:* (bottom right)
*Thread Reply:* Ahh I see - thanks. And will that # work everywhere? So it’s a way of linking pull requests on the same paper together?
*Thread Reply:* Yes, you can have multiple PRs to one issue, or multiple issues to one PR
*Thread Reply:* Got it 🙂 thanks again
@channel I've been informed by my supervisor that their review paper was 'accelerated' and they already reviews back.
Therefore: if we want to get a preprint out they can reference (and get 'CV worthy' citations), we should really try and get all the current singlegenome-hostassocaited papers into the list.
We current have just over 40 papers still to add. This means for everyone who is a part of this channel pitched in, that would just be 2 papers each (about an 1-2h in total worth of time for an average paper).
I would be very grateful if people could assign themselves to issues papers and make their PRs in ASAP! Remember: everyone who has made a contribution (as listed on github) will be a coauthor!
Ideally end of next week, I can maybe stretch it to end of first week of September
*Thread Reply:* I will absolutely be in by the end of next week--life calms down after Monday! Thanks for the timeline update!
did you have a conclusion on the consenssu/raw stuff in the end?
*Thread Reply:* Yes, I think so (@aidanva) Have a look at the single-genome slack channel 😉
*Thread Reply:* May I enter the holy grounds, oh god(dess)?
@channel Ele (@Ele) has made a wonderful set of (detailed!) step-by-step notes of going through making your own PR! So if you rather read prose than the slides I originally made, or still have questions after the slides, I would highly recommend reading through the guide:
https://github.com/SPAAM-workshop/AncientMetagenomeDir/wiki/Adding-a-Publication:-Step-by-Step-Guide
And importantly, as with everything here, you are welcome to make corrections/changes/additions to the notes! So if anything is unclear or you learn something else that is not already on there, feel free to add to it!
*Thread Reply:* Yes please do add to them - I am bound to have missed something!
*Thread Reply:* This is incredible!! Thank you Ele!!
*Thread Reply:* @Shreya — thank you! I hope it’s helpful 😄
Secondly, I had a thought last night, about the publication we will make. What do people think about trying to publish in Scientific Data?
My current justifications: 1) explicitly designed for publisihing this sort of thing (as in the name); 2) has OA publications and is from this month covered by the MPG central library (so no costs to specific PIs/institutes, I believe) 3) allows preprints 4) I wonder if we could fast-track review if we push to have it released alongside the review that inspired the creation of this dataset (which is also coming under the Nature 'family')
Of course I am open to other places to publish, however my only requirement is that they must be OA (and we have to be able to fund it somehow). Please propose them here for disucssion!
@channel Ash pointed out my guidance wasn't clear in the list READMEs regarding the BP dates. I will go back and check everything, but in the future - please calculate BP from 1950 not 2000. I apologise for my prehistoric bias (where +- 50 years doesn't mean much). I have updated the READMEs accordingly
@ivelsko Do you think the a b notation could be confusing if we don't update the key of the original publicatio
Do you think addng a .2, .3 etc. notation would be clearer?
Or how could we make it so that we don't have to go back and fix previous keys 🤔
I thought that keeping the 1st as-is, and adding a to the second, etc was fine. What’s not working about it?
Dunno I'm worried people might assume if there is a a that that is the first one
Could we now (before it gets too big) just add a , b as needed in one commit?
Doesn't solve future additions...
Or I could just allow from b onwards?
Ok, then better to set as b then
Merged straight ot master as it was pretty quic
If a same author/year combination already exists, please append a single lower case character (b,c,d etc.) to the key.
The already existing key does not need to be updated. b indicates the 'second' key added.
e.g. Muhlemann2018 (original), Muhlemann2018b (first duplicate), Muhlemann2018c (second duplicate) etc.
@channel reminder to please don't 'over-assign' yourself to issues. Only assign yourself to papers you are actively working on, to make sure there is enough to go around 😉
To be clear, really happy to see the enthusiasm! It is also helps me to keep track of what/when I can expect each issue to be done before our first release 💪
Also, Nik has kindly added AD to BP conversions on his tool! https://twitter.com/NiklasHausmann/status/1296741397882703874
*Thread Reply:* Note page updated too 👍
@channel I've just come across a case where two separate papres/teams published data from the same site (from C16th mummies from Italy. One was a single genome of HBV and another microbiome samples). However in the two PRs (on the different lsits) report the site names slightly differently.
How do you feel about also standardising site names in a restricted list (which would need to be updated at every PR...)? Or do you think this would then be becoming too complicated? Or would it be better to give guidance for people making PRs to check the different lists first?
I think it would be better to stick to the site name as it’s written in each publication, to avoid adding to the confusion
Could do two entries per site separated by a ; ( one for each publication) or have a notes column where you can write which other paper produced data from the same individual.
I think that's a bit ugly because then it's harder to filter that column when using the data (tidydata FTW). Notes isnormally bad as then it's not going to have the same consistent data and and clutter the table/data.
@channel reminder, as I made this my mistake (thanks to @Åshild (Ash) for poinitng this out; I'm going back and correcting everything now): we prefer uncalibrated dates C14 over calibrated. As the latter isn't always reported and calibration will change (see the latest IntCal2020 curve!), it's safer to use the uncalibrated atm.
I know that's my bad (I was reviewing with calibrated in mind, but Ash rightly said that's not what I had written :face_palm:). That said another thing I want to do at some point is go and collect all the Radiocarbon lab codes (as these are very hihgly standardised) so we can then get exact values for calibrating everyhting togheter 💪
I can help with finding the uncal dates, just let me know
It's going pretty quck getting uncal, so should be fine thank you!
Hi folks,
Ardelean2020 has samples from a sequence of strata. Not all strata have been dated, but most of them have. Some samples come from undated strata.
How reasonable would it be to estimate the date of undated-strata samples by taking the midpoint of surrounding dated strata?
For context: this is for sediment samples
Which would make me lean towards allowing it as they are presumably a little more secure than bones etc (depending on bioturbation)
@Becky Cribdon and @James Fellows Yates: Would not recommend interpolating 14C ages from cave strata (unless very well constrained), as cave sedimentation rate is often very uneven and can be reworked.
This is also an issue for lake sediment records (which usually have an undisturbed and reasonably constant sedimentation rate), where there are often few 14C dates and the ages of depths (= samples) are determined using a model of calibrated age to depth.
Better instead to also include the published calibrated age, but with an additional column to state which calibration curve was used?
(again with the assumption it's a bulk block, not saying the sediment is completley static)
OK fair point. I would say then @Becky Cribdon to just drop those samples.
I'm currently against the calibrated dates because calibration curves change over time. As I said above, in the future we can go back and pull all the exact radiocarbon lab codes, dates and error margins to allow people to do updated calibration (e.g, with IntCal2020)
(unless only calibrated dates are reported)
For samples within the last 100 years, should we still round up to the nearest century? as if something is has a BP date of 6, should then be rounded to 0?
well... that's another discussion, basically I am talking about the malaria genomes from 1944 from spain
then witihn ;ast 100 years == 100
Yes, 60 years is ancient
what is the cut-off again? more than 20 years? 10 years?
(And I swear there is a paper that says that but Ican't find it 😆 ).
no, when we get ot htat point it should be wihtin the last 100 years
Otherwise people will get the wrong idea
ok, then I will modify it, I can add the line in the README then
actually is already there but the example is from something 50 years old, if you guys agree I will add another example saying anything between 10 and 100 should be round up to 100?
but we should be careful how its phrased. Suddenly all samples pre-2010 become ancient
which will be problematic for samples between 1951-2010...
ok,so if a sample is from 1960, we don’t consider it ancient?
maybe we should say pre-2000 and from museum/archival material is ancient? thinking out loud here
(Aida can you add that to all READMEs pleasE)
so I will write something like: For samples pre-2000 and until 1900, the date should be indicated as 100?
that will also include samples up until 1850 if you do the calc based on 1950
Say anything more recent than 1850 is 1000
or we start using minus years, but that will add a whole other world of confusion
1850-2000 == 100
KISS (to quote Antonio, to quote othe rpeople)
sample_age In Before Present (BP) format i.e. since 1950 AD • When in doubt: https://nikhausmann.shinyapps.io/BP_to_BC_and_more/ Single date rounded to nearest century (i.e. end in '00') • For samples more recent than 1850, the age should be assigned as 100 • e.g. something only 50 years old would be assigned as 100
is the uncalibrated dat prferred still there?
since some people (not me 🙄 ) only read the first lines of the description
PLease make sure they are all synced across eahc list though ofc
(need to think of a better way for that 🤔 )
Not having to update four similar but slightly different READMEs
For reference (@Ele maybe you could keep an eye on these conversations and update your wiki as needed):
[13:37] aidanva what shall I do when I have country of isolation but not coordinates?
[13:37] James Fellows Yates Middle of country but no decimals
Thanks @Miriam Bravo! For adding Kerudin! It's now merged. I had to fix something else in the backend which is why the PR gt a little messy during merging 😅
I apologize for that, I got a little bit confused
No no, it was all me, you did everything right. We added a new column in parlalel to you making that PR and I needed to add it back in for you but I made a mistake while oding it 😆 so had to fix it
Hey all! We have decided that for the first release of ancientmetagenomedir we will only include papers with data prior to the 1950s. After the first release, we can then add samples that are younger. @James Fellows Yates will tag all the papers that fullfill this criteria and people can assign themselves to those
Also, there is a new column under the singlegenome-hostassociated called 'genome_type' which indicates whether the reported genome is a whole chromosome or an 'organelle' (e.g. with eukaryotic pathogens and their mitochondria, which may still be useful)
We also need some release names (e.g. nf-core/eager uses Medieval Baden-Württemberg city states; nf-core/sarek uses national parks of Sweden, other software uses a adjective + noun system e.g. copper sparrow)
Maybe cultural sites on the Unesco Wolrd Hertiage list?
Yes, we can use famous archaeology sites from the list
@channel 'milestones' for release tracking can be seen here: https://github.com/SPAAM-workshop/AncientMetagenomeDir/milestone/1
So please assign yourselves/work on those ☝️
Release v1.0.0: Ancient Ksour of Ouadane Due by September 04, 2020 Last updated less than a minute ago Release Description First major release of AncientMetagenomeDir of Host associated meta- and single-genomes samples older than 1950. Ancient Ksour of Ouadane, Chinguetti, Tichitt and Oualata: Founded in the 11th and 12th centuries to serve the caravans crossing the Sahara, these trading and religious centres became focal points of Islamic culture. They have managed to preserve an urban fabric that evolved between the 12th and 16th centuries. Typically, houses with patios crowd along narrow streets around a mosque with a square minaret. They illustrate a traditional way of life centred on the nomadic culture of the people of the western Sahara. Description is available under license CC-BY-SA IGO 3.0 from https://whc.unesco.org/en/list/750/

@channel @Maxime Borry has made a much imporved error reporting: (one for @Ele and her Wiki page!)
And now even better: the robot overlords have arriveD:
Every new commit on a pull request will see validation checks run. If an error occurs, a github bot will post the error for you
Though the annoying side effect, is that it doesn’t appear in the log anymore https://github.com/SPAAM-workshop/AncientMetagenomeDir/runs/1031512588?check_suite_focus=true
*Thread Reply:* And no artifacts are saved
Maybe you could also cat the content of each validation step, so that it still appears in the log ?
*Thread Reply:* Replacing it with | tee -a
*Thread Reply:* But not sure what's wrong with the artefact uploadin. I changed it slightly but maybe calling it wrong
*Thread Reply:* @Maxime Borry artefact is back, and comment always runs, but using | tee removes the fail tick 😕
*Thread Reply:* And also hides the error, which makes the check pass 😉 We don’t want that
*Thread Reply:* yeah that's what I meant
*Thread Reply:* Have an idea, one moment
*Thread Reply:* $ set -o pipefail
@James Fellows Yates So I nearly got my entry done for Martin et al. 2013, but their project accession (PRJEB0415) seems to have been deleted from the SRA and I can’t the data anywhere else. I’ve written to the authors for help. Of course I get the dataset with “problems” ;)
*Thread Reply:* @Christina Warinner https://www.ncbi.nlm.nih.gov/bioproject/PRJEB4015
*Thread Reply:* Reverse searching found it
*Thread Reply:* No idea what the original search was doing
*Thread Reply:* I found it - there is a typo in the accession number in the original paper
*Thread Reply:* Okay, silly question. Now I want to add in this data. How do I overwrite these entries? Do I just make a new pull request?
*Thread Reply:* Oh wait - I can see that I can edit my previous pull request. Nevermind!
*Thread Reply:* Okay - I think I did it! 🎉:mask_parrot:
*Thread Reply:* I might need to add those species to the list of valid species but if no other errors are reported then it's ready for review
*Thread Reply:* I think I fixed it and now the checks are running
*Thread Reply:* Nope, the checks failed again. It keeps failing on test ancient single microbial genomes
*Thread Reply:* OK, one sec
*Thread Reply:* Might be the webpage it takes the valid lists from hadn't updated
*Thread Reply:* So my pull request has never passed checks, so I just continue to mark it as new rather than a correction, correct?
*Thread Reply:* Oh my bad, I updated the wrong list
*Thread Reply:* No problem! It passed! Now I just need a reviewer 😉
*Thread Reply:* Mmm, no that was a different PR that passed, where you just edited the enum list
*Thread Reply:* In th efuture I 'll put the docs so if you need ot add a new sample host or pathogen you ping one of the core team
*Thread Reply:* Because I realise this is a little complicated
*Thread Reply:* OK! Now tests have passed!
*Thread Reply:* Yes, please ping Ash, Me and Irina in the reviewer box!
*Thread Reply:* I don’t even know what that means!
*Thread Reply:* I think I may have done it
*Thread Reply:* Yes you have perfect 😄
*Thread Reply:* Thank you very much! I might be able to look a tit tonight
@channel FYI, after discussing with @Miriam Bravo, for host-metagenome we've decided to re-list the communitytype of any tooth samples where a study analysed the oral community to 'skeletalmaterial' and 'tooth' (instaead of 'oral' and 'tooth'). This is because this leads to inconsistencies between other papers that look at teeth but without an oral microbiome focus, and really the tooth mateiral is unlikely to have major amounts of oral taxa (vs dental calculus, which is exactly what D.C. is).
We can argue that if people want to use the dataset to get more genomes or oral taxa, that they can still try screening any 'oral cavity' related sample_type
even if the community_type is not directly listed as 'oral'
I have also started a 'complaints' wiki page where we can jot down thoughts/opinions on the difficulties you have had when adding a study to AncientMetagenomeDir in terms of how people report thigns. This will be a very useful page for SPAAM2 and later discussions on defininng minimal reporting standards for the field! So please to anyone who has made PRs, add your thoughts/comments, this would be very useful!!!
Anotherone for @Ele and her notes if a sample age is not reported they should not included. This particularly goes for sediment studies, e.g. taking a midpoint between two dated layers is not sufficient due to bioturbation
And one more: release numbers will be via Calendar Versioning (so release one will be: v20.09: Ancient Ksour of Ouadane), as this is better for regular data releases
And more me (sorry for the SPAAM today ;)).
I'm going to start drafting the paper, please see the following document for my current thoughts on an outline:https://docs.google.com/document/d/1qButPlqSf4YZBv8pYyzyIvQ9fiPL-9YTdJuj4IshV4o/edit?usp=sharing
Thoughts/comments welcome!
I will share the actual draft once I've got a skeleton
(I'm doing this on Overleaf because they allow direct submission to the journal, no faffing with word formatting etc)
(thumbs up for Overleaf)
To view it while I'm drafting (read: word vomitting): https://www.overleaf.com/read/fjckjfvnqvmx
Then once it's mostly done I'll share the edit link for those who want to make direct comments (for those comfortable with LaTeX) and otherwise I'll share PDf/Word whatever for those who don't
OK, James rant:
Rule 1 of SPAAM guidelines/whatever we want to call it: Tables embedded in PDFs are BANNED 😡
flip table
10 to go until first release!
Dear @channel
I've just been informed that we have a soft deadline of end of Tuesday 1st September (CEST!), and a hard deadline of end of Thursday 3rd.
Therefore I would like make a release (to send to the authors of the review that the 'Dir was inspired from), and submit the preprint, ideally 1st of September and latest midday (CEST) of 2nd of September.
Therefore those with open PRs please let me know if you can make it or not (otherwise I will help out). Also, I will have the preprint finished by ~16:00 CEST (minus figures) tomorrow, so those who are listed as contributors (i.e. have had PRs merged into master), please keep an eye on the draft for you guys to skim read.
For those who still have have started their contributions (e.g. for list environmental/anthropogenic) but not yet merged. Don't worry: even if you don't make it into the first version of the preprint, we will wait for you guys to merge into Master before updating the preprint, and submitting to the journal. But I would like this to be no more than 2 weeks (so hard deadline 11th September).
Let me know if you have any questions! Sorry for the late urgency!
Cheers,
90% completed manuscript can be seen here for comments already: https://www.overleaf.com/read/fjckjfvnqvmx
To add comments on overleaf, press 'review' button in top right, then highight the line you want to comment on and press 'add comment'
But if you're not comfortable with playing with that, sending via email/slack (preferably to this channel!) is fine!
I don’t see a ‘review’ button on either version that appears when i click the link, do I need to look at something else? I tried viewing the left panel in Rich format but didn’t see it there, and still have only a ‘share’ button when I click the arrows for full screen
What about this one: https://www.overleaf.com/6976312443bkrvfhhfjvfq
*Thread Reply:* I finished making my comments for the pre-print version. Are you planning to include any figures or tables for the pre-print, or will you hold off for the publication b/c of the time crunch now?
*Thread Reply:* Will be generating a couple this afternoon (number of papers over time, and number of published samples) and @Åshild (Ash) is making a map.
If you have any other suggestions let me know!
*Thread Reply:* Have max of 3 objs though IIRC
*Thread Reply:* @Åshild (Ash) you’re not basing the map on the one Tina made, right? I found that one difficult to read
*Thread Reply:* It's awful isn't it 😆
*Thread Reply:* I can’t believe they’re going to publish it
*Thread Reply:* Is that the one you showed me James?
*Thread Reply:* then no
Anyone up for a quick review of a 2 sample paper?
https://github.com/SPAAM-workshop/AncientMetagenomeDir/pull/237
Only three more PRs to complete and ew can do our first release!
This is incredible! You have all had a busy week whilst I was away!!! I’ve updated the wiki with everything except the error reporting (will do that shortly). Do you want me to add some more data in? I have some time today 👍
Yes, we had a good surge last week 💪
I think we are good for now submissions, as all the papers have been assigned.
The priority would be getting your comments on: https://www.overleaf.com/6976312443bkrvfhhfjvfq

*Thread Reply:* Looks really awesome - thanks for putting all this together. I have added some comments (mainly suggestions to rephrase stuff)
*Thread Reply:* Oops sorry about the missing NHM affiliation, I think I accidently over-wrote when updating the 'leaderboard'. It is in the acknowledgmenets at least. Will re-add it now!
*Thread Reply:* Ahh it’s fine! You’ve got a lot on!!!! Thanks 🙂
Then there are a few open PRs (primarily environmental) that are looking for revies
@channel Are we going to include ancient shotgun RNA studies? for example: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000166
*Thread Reply:* How did they make cDNA? Did they use mammalian polyA tails? If so, then it’s host-specific and not metagenomic (unless you expect to find host-specific viruses in the data)
*Thread Reply:* They generated shotgun RNA data
*Thread Reply:* They also did some metagenomic analyses on the data to look for RNA viruses, but didn’t find anything
*Thread Reply:* I agree, I would leave out RNA for now
*Thread Reply:* But in the paper we have to state that we also have RNA pathogens in the dataset, and not just DNA
*Thread Reply:* But were the RNA viruses ones that integrate into host DNA and would have been DNA sequences?
*Thread Reply:* “Lastly, we looked for evidence of viral infection from RNA viruses (both single-stranded RNA [ssRNA] and double-stranded RNA [dsRNA]) in all the sequenced tissues, noting that previous aRNA work has revealed RNA viral genomes in ancient material. We found no evidence of viral sequences in our RNA data.”
*Thread Reply:* I meant which RNA viruses are included in the database already?
*Thread Reply:* and Influenza A potentially, but we need to discuss it
*Thread Reply:* there will be more aRNA virus papers in the future anyway
*Thread Reply:* I checked the method used for amplification in that paper and in theory it would pick up microbial RNA if it were there
*Thread Reply:* I would still leave out the “total RNA” paper. If you have methods to validate RNA viruses in DNA extractions that’s a different matter. Ancient metatranscriptomics is still not well-validated in my opinion
*Thread Reply:* OK, then we leave it out. Can always add in a later release if that part of the field develops and becomes more validated
we already include an RNA virus (eg Measles morbillivirus)
@Ele @ivelsko I've added a few sentences that needs your review (that it satisfies your comments)! I accidentally resolved (and should've re-opened) about introducing SPAAM, let me know if you can't see the comment...
You might be able to work (1) into (2)
*Thread Reply:* Ok, will give it a go
(shitty draft), but something like this?
I am out now, will send you later some feedback
I honestly would prefer to have a table. I'm not sure how much the workflow actually helps
*Thread Reply:* I think both would work. The workflow is basically a graphical abstract in this case. If there’s space for two it makes sense for a table to go in that specifically details what’s being recorded. The figure you sent looks to achieve this 🙂
Then can have richer info (like @Antonio Fernandez-Guerra suggested elsewhere, with technical info about validation)
Ok I've decided to split the figure above. @Antonio Fernandez-Guerra volunteered to use his graphic design skillz to make a good annotated workflow. I'll make a more detailed table with info about common columns
For those going through the draft, we are currently 88 words over the limit in hte Background and summary. Suggestions for removal of words/sentences/sections welcome!
I just shortened the abstract to fit within the limit!
*Thread Reply:* Oh, so then there are two options. this one is 162 words:
*Thread Reply:* Ancient DNA and RNA are valuable data sources for a wide range of disciplines. Within the field of microbial archaeology, the number of published genetic data sets has risen dramatically over the past few years, and tracking this data for re-use is particularly important for studies seeking to reconstruct the phylogenetic histories of pathogen lineages and studies that aim to explain ecological and evolutionary changes in microbial communities through time. AncientMetagenomeDir (archived at \href{https://doi.org/10.5281/zenodo.3980834}{https://doi.org/10.5281/zenodo.3980834}) is an index of published genetic data deriving from ancient microbial samples that provides basic, standardised metadata and accession numbers for rapid data retrieval from online repositories. This list is community-curated and spans multiple sub-disciplines in order to ensure adequate breadth and consensus in metadata definitions, as well as longevity of the database. Internal guidelines and automated checks to facilitate compatibility with established sequence-read archives and term-ontologies ensure consistency and interoperability for future meta-analyses. This collection will also assist in standardising metadata reporting for future ancient metagenomic studies.
*Thread Reply:* And this one is 148: Ancient DNA and RNA are valuable data sources for a wide range of disciplines. Within the field of microbial archaeology, the number of published genetic data sets has risen dramatically in recent years, and tracking this data for re-use is particularly important for large-scale ecological and evolutionary studies of both individual microbial taxa and microbial communities. AncientMetagenomeDir (archived at \href{https://doi.org/10.5281/zenodo.3980834}{https://doi.org/10.5281/zenodo.3980834}) is an index of published genetic data deriving from ancient microbial samples that provides basic, standardised metadata and accession numbers for rapid data retrieval from online repositories. This list is community-curated and spans multiple sub-disciplines in order to ensure adequate breadth and consensus in metadata definitions, as well as longevity of the database. Internal guidelines and automated checks to facilitate compatibility with established sequence-read archives and term-ontologies ensure consistency and interoperability for future meta-analyses. This collection will also assist in standardising metadata reporting for future ancient metagenomic studies.
*Thread Reply:* Mmm problem is the environmental stuff isn't looking at microbes necessarily, they are also looking at wider ecological profile (animals and plants)
*Thread Reply:* Can change microbial archaeology to ancient metagenomics
*Thread Reply:* Yeah, I think the second is better as it's less focused on microbes
*Thread Reply:* (so easier to update)
*Thread Reply:* Okay, how about this: Ancient DNA and RNA are valuable data sources for a wide range of disciplines. Within the field of ancient metagenomics, the number of published genetic data sets has risen dramatically in recent years, and tracking this data for re-use is particularly important for large-scale ecological and evolutionary studies of individual microbial taxa, microbial communities, and metagenomic assemblages. AncientMetagenomeDir (archived at \href{https://doi.org/10.5281/zenodo.3980834}{https://doi.org/10.5281/zenodo.3980834}) is an index of published genetic data deriving from ancient microbial samples that provides basic, standardised metadata and accession numbers for rapid data retrieval from online repositories. This list is community-curated and spans multiple sub-disciplines in order to ensure adequate breadth and consensus in metadata definitions, as well as longevity of the database. Internal guidelines and automated checks to facilitate compatibility with established sequence-read archives and term-ontologies ensure consistency and interoperability for future meta-analyses. This collection will also assist in standardising metadata reporting for future ancient metagenomic studies.
*Thread Reply:* Sounds good to me! Thanks!
*Thread Reply:* You can update the file directly
*Thread Reply:* I am, but just wanted to run this by you too
Background/summary is the bigger issue I think
*Thread Reply:* Okay, all done with editing and commenting!
*Thread Reply:* Intro is now just 28 words over
*Thread Reply:* Thank you very much!
With @James Fellows Yates we have been drafting the following figure for the workflow. Please send us feedback and we will modify it accordingly :-)
*Thread Reply:* Very nice @Antonio Fernandez-Guerra!
it needs some polishing, but the main ideas are there
I'm not familiar with this format of figure, but is the box containing the block of code supposed to be connected to both "Metadata extraction" and "Schema validation"?
AS the contributor should normally be following those guidlines anyway when submitting (and then it's checkeD)
My only question is where it says “Is the same age Before Present….“, should the word “same” be there? Or should it be “Is the age Before Present…”
*Thread Reply:* Oops yeah 😅 my bad
@channel I've now added a basic map to the pre-print draft, so just the finalised workflow and we can post it on teh bioRxiv!
*Thread Reply:* Great job, @James Fellows Yates!
*Thread Reply:* It’s so much nicer than the one for the big review
*Thread Reply:* Doesn't have the time aspect though
*Thread Reply:* But you and Ash will work out how to add it in a visually appealing way for the publication
*Thread Reply:* @Åshild (Ash);)
*Thread Reply:* @ivelsko if you are map wizard, please go ahead and work your magic 😉 I will be using the data to test my map making skills, so won’ be the fastest
*Thread Reply:* No no, I just really really didn’t like the map Tina showed us
📣 v20.09 Ancient Ksour of Ouadane is RELEASED 📣
https://github.com/SPAAM-workshop/AncientMetagenomeDir/releases/tag/v20.09
DOI: 10.5281/zenodo.4011585
Thank you and much love and apprciated to everyone who has contributed, @channel! Pre-print will be finalised and submitted to BioRxiv this afternoon!
*Thread Reply:* Why is the version called “Ancient Ksour of Ouadane”?
*Thread Reply:* Because it's recommend to give text 'names' too, so I randomly picked stuff from the WHO Cultural Heritage list
*Thread Reply:* To keep it relevent
*Thread Reply:* Oh crap misspelling 😆
*Thread Reply:* Oh, and single genomes are missing
*Thread Reply:* In the release?
*Thread Reply:* Yes, the zip file doesn’t have single genomes
*Thread Reply:* that's odd..
*Thread Reply:* Oh found the issue, here comes the first point release 😆
*Thread Reply:* Can you send me the new link when it’s ready?
*Thread Reply:* Will do!
*Thread Reply:* Let me know if there are any other issues :face_palm:
And remember! This isn't the end! Please keep submitting issues/PRs, this is only phase 1 😈
Awesome! Thanks for spearheading this James 🙂 Pretty cool that a pre-print paper was pulled together in about 1 month
All thanks to you guys (and your tolerance of my pestering)
Slightly improved version. If you have any comments let us know, and we will do the last edits before adding it to the draft
(we wait for comments for the next 30m or so - but we can always update between pre-print and submission!)
@channel ☝️comments requested
I find that the “Metadata extraction” box pointing to the box with detailed validation scheme suggests some kind of automatic metadata extraction. Perhaps changing the text to something like “manual metadata entry” and removing the right-side arrow would clarify it. That way only the “Schema validation” box points to the code
both are using the schema, you are extracting the data based on the schema constrains. What we can add is something that claifies that the BRANCH and PR are user mediated
this is why we added a different color for this step
to indicate what is manually done and what it is done by authomatic check
maybe a bar on the left where you have the workflow separating between the role of users, volunteers, reviewers
then we show that there are three different points of validation
nice idea, fill up some the white space with a legend
If you can can provide some terms to use for: • the ones who suggest a paper • the ones that mine the paper • the automatic checks • review by curator
my vacation brain is not very imaginative for this 😅
@ivelsko? My english is failing me 😆
It's something like:
Proposer Contributor Auto-Validation Curator (although technically it's the same as a contributor, anyone can review)
*Thread Reply:* Those are fine. Proposer sounds a bit odd, but I cant think of another option
I would add curator, sounds more professional
*Thread Reply:* These are roles, rather than people, so including curator is OK. (in theory, one person could do all steps, aside from review)
will quickly add this and see how it looks
📣 Point Release v20.09.1 Ancient Ksour of Ouadane is RELEASED 📣
Because I'm a dumdum and missed one of the files in the ZIP file 😆
https://github.com/SPAAM-workshop/AncientMetagenomeDir/releases/tag/v20.09.1
Thanks to @Christina Warinner for pointing it out
Also in the future, I've written docs how to do a release for those interested/do it in the future: https://github.com/SPAAM-workshop/AncientMetagenomeDir/wiki/Release-Procedure
Potential solution, don’t look at the color though…
Mmm, that makes it very busy. I would rather have a legend for each thing and have a bar more like:
the problem is that it will be too colorful
I will make the labels less bold and streamline them a bit so they are cleaner
What about going back to the icon ideas
we can find icons for each role
Propose can be the double person.
Contributor a single person.
Curator the 'eye' icon
Auto validation is a computer
(or curator could be bodyarmor one 😆 )
I think as @aidanva says that would be cleaer and be less busy
in terms of human vs computer role
I prefer not to overload the figure
the facet strips doesn’t convince me too much
Stick with colours and legend you mean?
no, the strip where the facet titles are
*Thread Reply:* Ah I see. I decided to stick with theme_classic because I found it became a bit messy in the other figures (the timeline ones)
*Thread Reply:* i would check how it looks like without the top and side of the rectangle
*Thread Reply:* Looking if I can do that
*Thread Reply:* or add very very light grey for the background
*Thread Reply:* in the main plot
*Thread Reply:* so you have each facet delimited without the need of adding the y-axes and then a minimal facet strip
*Thread Reply:* @aidanva @Antonio Fernandez-Guerra
*Thread Reply:* I can remove the strip titles
*Thread Reply:* Actually no, I don't like this as getting the legend to not be clipped sucks
I think I will like to have the histograms on the right of the maps they correspond to
*Thread Reply:* Eek, ok. Might make it very wide but let me try
*Thread Reply:* yeah it mad eit too wide
*Thread Reply:* And caused the legend to be cut off
*Thread Reply:* I liked it more that way though
*Thread Reply:* but ok, I will remove the legend for the colours
*Thread Reply:* ignore me, since you are going for the long version my comments have no sense
I like having the labels still to help with colour-blindness
that goes from -180,180
Thanks so much for letting me be involved! So exciting to see it released!
Actually, I'm going to drop the histograms, getting the bins right is too difficult due to the long tails due to the sediments
*Thread Reply:* The auto-validation is out of line in the legend
*Thread Reply:* yep, I would like to have suggestions where to place the legend, not 100% convinced
*Thread Reply:* I think position is OK actually
*Thread Reply:* You could put a box around it though
I preferred the integration of the roles using the 'tab' system, rather than the line-'brackets', the lines are getting very busy there, unless you move roles to the right-handside of the time line
instead of the brackets I can just put the line next to the big arrow
I think that's fine for now
ok, let me export the pdf and png
Use this ones better, the dark grey text in the boxes looked bad
Starting bioRxiv submission now!
After 21039210302193 million minor formatting mistakes
```MS ID#: BIORXIV/2020/279570 MS TITLE: Community-curated and standardised metadata of published ancient metagenomic samples with AncientMetagenomeDir
Dear James A. Fellows Yates;
This is an automatic message acknowledging your online submission to bioRxiv.
Completion of article screening typically completes within 48 hours, however, it may take longer over a weekend or holiday.
If you would like to make changes to your submission prior to it being approved for posting on bioRxiv, please contact bioRxiv at biorxiv@cshl.edu and we will return your submission. Do not create a new submission to update your manuscript as this will create a duplicate submission and will significantly delay the screening of your manuscript.
In addition, note that bioRxiv now allows you to save time submitting to certain journals or review services by transmitting your manuscript files and metadata directly. To submit your paper to a journal, please click on the link below to access your "Submit bioRxiv Preprint to a Journal or Peer Review" queue: (https://submit.biorxiv.org/submission/queue?queueName=send_paper_away_author)
Thank you for your submission.
Best wishes, The bioRxiv team```
@Christina Warinner hopefully it will get past screening faster than 48h :face_palm: they've increased that (used to be 24h). Please let lead author of review know that citeable bioRxiv DOI is comiing
And @channel please reflect on it, if you think of anything to add/remove in the next 2 weeks we can modify it before submission to Scientfic Data
https://www.biorxiv.org/content/10.1101/2020.09.02.279570v1 @channel preprint is out!!!

@channel thank you again 😄
Now I would to ask you again for a little more of your time/expertise. I will be very briefly showing AncientMetagenomeDir at SPAAM2, but we want to focus on the challenges we have in scraping metadata, with the eventual goal of making a group to develop a standard metadata reporting guidline/table.
I need starting points of discussions, to help us triage priority problems. PLease can you add to the following Wiki page your thoughts/feelings on the problems you had when trying to scrape metadata for AncientMetagenomeDir; this will help me a lot make the presentation for discussion!
I'm going throught the preprint again and I had a thought - @Pete Heintzman pointed out we should say why we don't inlcude negative controls. I think it's fine to exclude them as this 'experiment' level data rather than sampling info; however finding these is important (if they are even uploaded 😏 ) - to help this though we could also include the PRJ-like project accession codes in addition to the actual sample SRS codes.
What do you think @channel? This should be relatively easily retrievable for everything that is already there programatically so we don't have to have everyone go back and find them all.
*Thread Reply:* I agree! I think that’s useful info to have anyway… these databases can be a little daunting for newbies so more info can’t hurt
*Thread Reply:* By negative controls I assume you mean samples that are chosen to act as negative controls rather than laboratory neg controls (extraction and library blanks), right?
*Thread Reply:* I've not seen really the former
*Thread Reply:* I had a soil sample in the salmonella paper that we used as an environmental neg control. It should probably be added
*Thread Reply:* Mmm for the list though we only report retrievable genomes
*Thread Reply:* I might be remembering incorrectly, but doesn’t is say somewhere on ENA/SRA that they don’t want lab blanks to be uploaded? Maybe I’m completely wrong in thinking this
*Thread Reply:* A lot of papers don't report negative samples so might not be worth it.
*Thread Reply:* Anyway, I think it’s a good idea to include them, if the data is available
*Thread Reply:* Mmm, I've never seen that. I asked them about which taxon to put when uploading my blanks and they gave me a hacky workaround that exists and said they are trying to improve that.
*Thread Reply:* > Anyway, I think it's a good idea to include them, if the data is available
Eek, this might be a bit late now. But honestly I think the same thing goes also for when looking data itself, you should still check the publication's themselves for all data and metadata
*Thread Reply:* The Dir should just act as a pointer to relevant papers, ultimately 😅
*Thread Reply:* Ok 👍 could consider adding a column later where is says if blanks were presented or not, and/or submitted to ENA/SRA. But this would be mostly relevant if we decide to comment on quality in the future
*Thread Reply:* My idea would be that with a future ERR/SRR list we would have a 'blank' category, indeed
I think is a good idea as well, also for our own curation, i.e having a similar check like the one we do with the schemas to validate that the sample ids belong to the project
*Thread Reply:* One for @Maxime Borry , which should work this time 😂
Also @channel - I've pre-started the submission (which I will make NEXT friday). The submission form still requires a 'title' for some archaic reason. For those non-Drs or non-Professors, I have put Mr./Ms./Mx, corresponding to what I assume your name implies if there is NO indication of pronoun on twitter (if you're on twitter).
If you have an alternative preference not listed, please DM me.
Just saying in case you get some correspondence from the Nature publisher
Any suggestions for potential reviewers?
I was currently thinking something along the lines of:
- Simon Rasmussen
- Tanvi Honap
- Vivianne Slon
- Raphael Eisenhofer
@channel reminder that I need everyone to confirm they've read the paper one more time and left comments by this Thursday (17th!) I will submit on the friday! Thanks to everyone who already has! At the bare minimum check name/affiliation, contribution and funding stuff is correct
If I don't hear back from you I assume you have no comments.
Thank you very much!
@channel Editorial board members suggestions also recommended! As to cover each sub-field
https://www.nature.com/sdata/about/editorial-board
Simon Ramussen is one, Philip Hugenholtz as well
Does anyone know about Kurt Kjær for sedaDNA?
I would have conflicts of interest with Kurt
I'm currently down to:
```Simon Rasmussen Technical University of Copenhagen, Denmark Expertise: Bioinformatics, Metagenomics, Ancient DNA, Next-generation sequencing, Genomics
Guojie Zhang University of Copenhagen, Denmark Expertise: Comparative genomics, Evolutionary biology, Functional genomics, Environmental genomics, Phylogenomics
Ben Marwick University of Washington, USA Expertise: Archaeology, Human ecology, Palaeoclimates, Text analysis```
All have aDNA/archaeology experience. Well Guojie not so directly
But the environmetnal aspect I don't know 😞
Even though Philip Hugenholtz doean’t have aDNA experience, I still think he might be a good choice
oh no wait, Ash, you published with Simon?
Yes, and am about to publish with Guojie, now that I come to think of it
Alternative reviewer (also for Environmental peeps: Mike Bunche?)
But I have no direct on-going collaborations with them (the paper with Guojie comes out on Oct 5th)
But I assume others on the list might have ongoing collabs with Simon
Do you think it's OK to put Simon as an editor (rather than reviewer) but note a possible C. of Interest?
What abot Carles Lalueza-Fox for pathogen/aDNA?
Yes, I thought you were listing editors? I’m confused
Sorry, - yes I also then realised Tanvi (reviewer suggestion) has also published with Tina
Side note about Viviane, she has her own wiki page! https://en.wikipedia.org/wiki/Viviane_Slon

I was recently asked to a review a paper and told that I potentially have a conflict of interest, and the editor told me that it is merely impossible to find reviewers in ancient DNA that haven’t collaborated with each other in recent years. As long as we reveal it, I think we can put some of them.
Especially for this paper since we are from so many different labs
@Åshild (Ash) do you have any suggestions for pathogen people?
I have one microbiome, one environmental now
I’ve also published with Susanna, but no ongoing stuff atm
good one, might be a bit better than Kelly
Ah but Jess is at McMaster
*Thread Reply:* Do you know where she still is?
*Thread Reply:* Ah she's back in Germany
*Thread Reply:* Yes, at the Charite in Berlin I think
There is also Hernan Burbano for plant pathogens and general aDNA
I was on a paper with him (the annual reviews one, same for Clemens)
Editors:
• Simon Rasmussen
◦ University of Copenhagen, Denmark
◦ Expertise: Bioinformatics, Metagenomics, Ancient DNA, Next-generation sequencing, Genomics
◦ Note possible conflict of interest: author Åshild Vågene is listed as co-author on a recent publication with Dr. Rasmussen (Jensen et al. 2019 Nat. Comms. doi:10.1038/s41467-019-13549-9)
• Ben Marwick (University of Washington, USA)
◦ Expertise: Archaeology, Human ecology, Palaeoclimates, Text analysis
Reviewers:
• Raphael Eisenhofer Philipona (<a href="mailto:raphael.eisenhoferphilipona@adelaide.edu.au">raphael.eisenhoferphilipona@adelaide.edu.au</a>, School of Biological Sciences, University of Adelaide)
◦ Expertise: Ancient DNA, Microbial Ecology, Microbiology, Conservation Biology
• Meriam Guellil (<a href="mailto:meriam.guellil@ut.ee">meriam.guellil@ut.ee</a>, Institute of Genomics, Universtity of Tartu)
◦ Expertise: ancient DNA, pathogen genomics, bacterial genomics, ancient microbiomes
• Barbara Mühlemann (<a href="mailto:barbara.muehlemann@charite.de">barbara.muehlemann@charite.de</a>, Charité – Universitätsmedizin Berlin)
◦ Expertise: ancient DNA, pathogen genomics, viral genomics,
• Michael Bunce (<a href="mailto:michael.bunce@curtin.edu.au">michael.bunce@curtin.edu.au</a>, Trace and Environmental DNA (TrEnD) laboratory, Curtin University)
◦ ancient DNA, Environmental Science, Biomonitoring, metabarcoding, environmental DNA
re. editors: • I too have a COI with Simon Rasmussen (although it is 6 yrs old): (Rasmussen et al. 2014 Nature. doi:10.1038/nature13025). re. reviewers for sedaDNA: • Mike Bunce is no longer at Curtin, but works now at the NZ EPA, so not sure if he still does peer review work. • Mikkel Pedersen and/or Beth Shapiro as an alternative? (I am sure there are loads of strict COIs with both of these options).
Ok, Mike is still listed on the TrEnD lab, so maybe he could select someone in his lab (from the press-releases it doesn't look like he's stopped working there necessarily)
MIkkel was also on the same paper as Simon Rasmussen with me
I am sitting next to Mikkel
*Thread Reply:* I said hi to Mikkel this morning
*Thread Reply:* You're a friendly bunch. I'm sitting all alone 😆
*Thread Reply:* Not now though
*Thread Reply:* Would be kind of strang ehe is now in my living room 😅
*Thread Reply:* I said hi outside the museum on my way to the lab, socially distanced
*Thread Reply:* Given that you're all on the same papers all the time it seems like you do live together 😉
I think editors is not so bad, it's reviewers which might be be more important. The four reviewers above we don't seem to have CoIs atm
Thoughts? Tina is also Corresponding author so she is listed in the cover lettter, but she has NOT read yet!
(I will fix the colours later)
*Thread Reply:* It’s such a funky green! :partyparrot:
*Thread Reply:* It's fucking awful 😆 It's a really old template but the only one I could fnd
@channel for clarification, I have no idea how to write a good cover letter so any suggestions welcome 😅
I think it reads well, I only have a few comments 😊:
- This sentence “with the finding data for re-use or comparative analyses not being trivial” needs to be rephrased. How about “…making it sometimes difficult to find data for re-use and/or comparative analyses”? or smth along those lines
- This is written a bit weirdly, I think it’s a typo: “Suitable reviewers may the found in the fields...”
- Only Barbara’s email is blue, either all emails should be blue or black
*Thread Reply:* For number 1, I want to emphasise finding because it's in FAIR. I realise there was a missing word, does it still read wierd/
"While the field of palaeogenetics is recognized as a leader in making raw sequencing data available in public archives, deposition does not always follow FAIR principles with the finding of data for re-use or comparative analyses not being trivial."
*Thread Reply:* Better, can you think of another way to phrase “not being trivial”, how about “is not trivial” or “is not always trivial”. ‘Being’ reads a bit passively to me if I am nit-picking
*Thread Reply:* I am struggling to get it to work without 'being' because of grammar...
"ata must be balanced with the protection of precious cultural and natural heritage. While the field of palaeogenetics is recognized as a leader in making raw sequencing data available in public archives, deposition does not always follow FAIR principles, with the finding of relevant data for re-use often being difficult to do"
*Thread Reply:* "While the field of palaeogenetics is recognized as a leader in making raw sequencing data available in public archives, the finding of relevant data for re-use is often highly time consuming, as deposition does not always follow FAIR principles."
*Thread Reply:* I will change this sentence: “of which are decided upon through consensus of curators and ensure consistency through out the lists” to “of which are decided upon consensus of curators and ensure consistency throughout the lists”. You use the word through in that sentence and the one that follows 3 times
*Thread Reply:* Fixed it and replaced other 'throughs'
Final Map and sample age timeline (thanks to @Antonio Fernandez-Guerra for thta!)
If others agree could you change the legend for the sample ages so that the colour order matches the figure (blue on top, then pink, then green)? It helps to have the colour order matching the figure (for me at least), instead of alphabetical as it is now. But maybe that’s just an OCD thing I have …
+1 also you might want to try to remove the facet strips to see how it looks like
*Thread Reply:* It doessn't work for the other plots because they are too close to each other
*Thread Reply:* And so I would rather have it consistent
I wish I fucking could but sometimes my R skills fail me 😞
send me the code you have and I fix it
*Thread Reply:* I think the problem is that the factors don't work properly as you add three different bar plots
*Thread Reply:* I am in my phone now, but in https://github.com/SPAAM-community/AncientMetagenomeDir/blob/master/assets/analysis/ancientmetagenomedir-analysis-notebook.Rmd#L403 add breaks = names(dir_colours)
Couldn’t test it, but I believe it should do the job
*Thread Reply:* The values bit?
*Thread Reply:* you set the order of the legend keys by setting the breaks
*Thread Reply:* Didn't know that! Thanks!
the maps look very cool! so many metagenomes
you could have changed it in inkscape as well
re. panel B: any reason the x-axis only goes to 100,000 years? Several of Slon2017’s sedaDNA sites go back further than this (up to 550 kyr).
Oh good point, I need to add that to the caption.
Because otherwise everything gets massively squished and you can't see anything
Good catch @Pete Heintzman, thanks!
@channel final version of the manuscript has now been submitted to Scientific Data. Thank you once again for all your help and support and patience with all my nagging ❤️
@channel I'll be presenting the 'Dir in SPAAM2 today; you have about 2 hours to post your criticisms/issues/problems when getting metadata from publications, so I can include them!
Preferably here (so I can quickyl add them to the slides now), but also on the wiki: https://github.com/SPAAM-community/AncientMetagenomeDir/wiki/Issues-Encountered-when-Metadata-Scraping
• Tables that are images • Citations of citations, go down the rabbit hole to find the original description of the site • Inconsistent or no report of the site coordinates • Lack of tables reporting C14 dates, and particularly raw C14 dates --> does not allow for a recalibration of the dates • Rarely tables including the ENA/SRA codes
Yup, the C14 dates especially, I was surprised at how few report the raw uncalibrated dates
Also, the site can be really hard to find. Often the site name is in English in the paper and is therefore impossible to find on googlemaps where the site name/church name etc. Is in the original language
As @aidanva said, reporting of coordinates would be a huge help
I also agree that the coordinates would really help but a lot of archaeologists are not willing to reveal them in order to avoid others to find their exact excavation site, which is a pain.
it can always be approximate coordinates
Which I will argue would be better, since you also probably want to avoid people to go to archaeological sites and vandalising them
*Thread Reply:* One can always remove a decimal to obscure a site’s exact location
*Thread Reply:* yeah, that’s what I was proposing
This is a very good point. But if it is a known site, ie a church, then it would be super helpful if they could also specify the name in the original language/or coordinates to make it more ‘findable’
I wasn’t aware of this initiative before today but I think it’s a great idea. I’m wondering though why library treatment (nonUDG, halfUDG, fullUDG and capture/shotgun) are not part of the metadata? I think it is quite crucial to have this information, and sometimes it’s not that easy to collect, especially when papers include all kind of different treatments.
Samples != Libraries. I initially thought to du that at the same but that is a even more massivr clusterfuck
But watch this space ;). As we have the SRS codes we can make base templates in which makes easier for people to fill in library info. Sample level also gives us an idea of the scope of the work
Which imo at the moment might be feasible
Oh I see. But I think that would be one direction for future development?
*Thread Reply:* "watch this space"
@channel a couple of updates: AncientMetagenomeDir preprint is now officially in formal peer-review
And secondly, there was a question from one us and I would like to let you know my thoughts:
If you are scheduled to present either at a conference or workshop (or dept. meeting or w/e!), and struggling to find something to present because COVID I'm sure has affected a lot of people's data generation: please feel free to give presentation on AncientMetagenomeDir! Getting the word out would be really great, and of course this is a team/group- effort, and it will only continue to be successful if it stays like that. So ownership of the project is shared with everyone, and therefore you may present on it.
The only things I ask are the following:
1) You must inform/ask the whole group first (also useful to prevent multiple-presentations at the same event 😉 ) 2) You should not list yourself as the sole author in the abstract/presentation (if there are restrictions we can see if we can find ways around this)
- This is particularly important if there are some form of published proceedings or similar 3) The conference must allow re-presentation/publication at other conferences
- Also particularly important if there are some form of published proceedings or similar - others may want to present etc., and this may be an issue until the actual publication is out because the journal might not like this. 4) You make the slides available to the group before/after the conference (for comments and then for their records), and ideally we can upload the slides to the website or somewhere public.
Feel free to message me if you have any questions
Another (old) paper for team PathoPeepz: https://github.com/SPAAM-community/AncientMetagenomeDir/issues/285
Ancient viruses from Japanese teeth: Ancient viruses from Japanese teeth? (maybe one for @Maria Spyrou?)
https://github.com/SPAAM-community/AncientMetagenomeDir/issues/286
here comes a big 'un for microbiome/pathogen people! https://royalsocietypublishing.org/toc/rstb/2020/375/1812
@channel half of the papers are paywalled, could maybe someone with access add them as issues (if the data is avaliable, you can check that on the left hand side as there is an explicit header called 'Data Accessbility')
@Miriam Bravo I'm going to auto-assign you to your paper (https://github.com/SPAAM-community/AncientMetagenomeDir/issues/296) for obvious reasons, but you can un-assign yourself if yo udon't have time
Hi Everyone! I'm new to the group but have been lurking on the github for a while. I thought of a few opportunities where I might be of some help (Github Issues Link). Just wanted to get acquainted and caught up with all the amazing work in this group:
- SRA metadata. I've got a tool over at NCBImeta that pulls and organizes metadata from NCBI (bioproject accession, biosample info, SRA sequencing depth, etc.). If this tool isn't a good fit, I have a lot of experience working with NCBI's API and parsing their XML format...
- Databases are a passion of mine. Text files are amazing for wide-reaching accessibility, but if there's talk of scaling up, I could help with SQL work.
- Geocoding. I've got a project right now where I'm curating and 'standardizing' the geospatial information and geocoding it (programmitically with GeoPy). Cheers, and nice to meet everyone!
@channel what do you think? Have a look at the tool Katherine has made and we can think if we can integrate it somehow.
One thing that was requested a lot was getting direct links to FASTQ files, and their corresponding library information (e.g. what sequencer sequenced on, whether they were UDG treated or not etc.), so maybe we explot it for something like this?
*Thread Reply:* Does the community have a preference for where everyone gets their FASTQ files from (ex. NCBI, ENA, etc)?
*Thread Reply:* IIRC most uploads are to ENA (at least for microbiome) and I share a hatred with a few people of fastq-dump and the SRA file format
*Thread Reply:* But good question. We can make a poll
*Thread Reply:* Yes fastq-dump causes me a disproportionate amount of project frustration...
Also while I'm spewing ideas, I wanted to add an extra point:
- Web apps. I'm in the (slooow) process of converting my command-line tools to web applications. I was curious if there's been any desire to turn ancientmetagenomedir into a web app that external users can interact with.
Not perse, the furthest idea I had was displaying plotly summary images on the website.
But something like https://sra-explorer.info/ might be useful indeed?
You can also do a poll to see if people would be interested in that!
So it looks like it's pretty split 🤔
@Katherine Eaton, the main experience from us gathering the data for the 'Dir was that people upload metadata in a crazy amount of different ways, and that we can't really trust what is uploaded. Until metadata upload is standardised (see <#C01BX7EM4EL|metadata-standards> for future developments in this area), whatever is on SRA/ENA might not be so initially useful to perform automatically pull down because it'll have mistakes/inconsistencies etc.
What might be useful is if we could use your tool to take all the ENA/SRA sample IDs to generate lists of all the libraries and their related FASTQ files, which we could manually go back and curate to make sure it is as we expected. It would also help us to assess what metadata is already there.
My question for @channel would be, if you wanted to get metadata on libraries (and FASTQ files), what information would you want. Please reply to this in a thread!
*Thread Reply:* I would want to know:
- which FASTQ files are from the same library
- what damage treatment (if any) was used
- what library-build enzyme(s) was used
- what sequencer they were equenced on
- how many reads
*Thread Reply:* -read length -batches (extraction, library building, pooling, if different) -indexing strategy (are there potentially internal indexes that need to be removed?)
*Thread Reply:* -DNA molecules per uL of the library (not the pool, for decontam)
*Thread Reply:* • Library layout (paired-end, single-end)
*Thread Reply:* • (Basically all the fields needed for the tsv input of nf-core/eager 😃)
Maybe also: https://link.springer.com/article/10.1007/s00334-020-00805-y but I can't access it

*Thread Reply:* #notNGS
@channel just as a reminder, we still have ~20 issues still open https://github.com/SPAAM-community/AncientMetagenomeDir/issues. So if you have a spare 20 minutes would be great if you could assign yourself and add to the lists 🙂.
@Katherine Eaton is also now working on automating gathering some of the FASTQ level stuff (you asked for this @Marcel Keller, for example), to make it even more useful 💪
Reviews for the paper are back! We are now in editorial decision stage 🤞
@channel "I am forwarding your submission for minor revisions. "
🎉 you should all have had an email from the editor so you can see the reivisons
So task assignments (sorry for duplicates, I contacted a few people privately already):
@Abby Gancz is looking into seeing if it's possible to upload the Weyrich 2017 data to the ENA/SRA to make the editor happy the data is on a more 'recognised' repository
@Antonio Fernandez-Guerra will be updating the workflow figures for consistency with the text
@Ele do you think you could update the wiki screenshots to reflect the latest workflows (in particular the new auto-messages telling you if there is an issue or not - so you don't have to look at the wierd GitHub actions sub-pane)? Maybe you could do a submission of one of the open issues 😉
@Becky Cribdon @Pete Heintzman in particular see <#C018UBC9T47|dir-environmental> about whether we should add an extra column for the environmental stuff
Everyone else: I will be sending the new overleaf link probably beginning of next week for your thoughts on suggested changes!
*Thread Reply:* Hey, Yup should have some time over the next day or so 🙂
Thanks all again, in advance <3
@channel
Revised manuscript: https://sharelatex.gwdg.de/1575121292kpsgdpkzgrbb Cover letter:https://sharelatex.gwdg.de/4133753148fhprtsbwvnxh
Please note the follow
- this is a 'locally' hosted version of overleaf that allows track changes
- please do not accept/reject changes (so others can also see it), but please leave or modify the change comments if you are not happy with it (I'm sure @Becky Cribdon will find many problems with my english :P).
Please let me know once you've read the manuscript and you think the changes are sufficient or you have any major issues.
Feedback on the cover letter would be also welcome - in particular the section on the Radiocarbon dating!
EDIT: Please also leave your name in any comments you may leave!
*Thread Reply:* When do you need the comments by?
*Thread Reply:* Friday ideall
*Thread Reply:* (see email)
*Thread Reply:* Finished adding in my (very minor, mostly typo correction) edits in track changes. I noticed that Query 23 does not yet have a response - I’m happy with either option (keeping as is or making the terms the same). Nice job everyone, and especially @James Fellows Yates ! :mask_parrot:
*Thread Reply:* Thanks very much @Christina Warinner! For query 23 just waiting on Antonio to update the image with the new terms in the text :)
Hey all, been a while since I tackled an issue and I’m getting confused. Could someone help? I am looking at Morozova et al. 2020 who have assembled 4 Y. pestis genomes. I assume for the Dir we want these 4 genomes recorded right? When I go to the supplementary there are lots of human metagenomes and a few rat metagenomes and it’s thrown me off! Should I actually be logging these? TIA
*Thread Reply:* paper: https://royalsocietypublishing.org/doi/10.1098/rstb.2019.0569#d1e1863
*Thread Reply:* data: https://www.ebi.ac.uk/ena/browser/view/PRJEB35426
*Thread Reply:* Also won’t be available for the next hour or so - I promise I am not ignoring anyone and I’d be really grateful for any tips! 🙂
*Thread Reply:* What sort of metagenomes 🤔
*Thread Reply:* Do you mean they are just listed in the ENA as metagenomes? Or do they actually analyse in the paper the data as 'metagenome'
*Thread Reply:* Then just list them as single genomes
*Thread Reply:* Technically all aDNA are metagenomes which is why people put the but if they've not characterised the metagenome as a whole we can ignore
*Thread Reply:* Great - thank you! Sorry for the confusion.
*Thread Reply:* Yup they got a low coverage genome iirc
*Thread Reply:* Only list samples they yielded genomes from though
*Thread Reply:* Hey @James Fellows Yates another questions 🙄 This paper has uploaded 4 separate libraries for a sample. So the same sample has 4 different sample accessions. Which do I pick?
*Thread Reply:* All four, you can check other examples: we use a comma separated list
*Thread Reply:* In the wiki I have helpfully noted “Careful: You may have multiple ERS/SRS codes per sample as some people upload each library as a different ‘sample’. ” but not how to tackle it!
*Thread Reply:* ERRXXXX,ERRXXXXX,ERRXXXX
*Thread Reply:* Will update notes too!
*Thread Reply:* I'm late to the game, but @Ele , I struggled with the questions too (like James said..all aDNA are technically metagenomes..so I was documenting everything. woops)!
Having trouble with some checks failing because of enums, I have updated the lists on my branch, does the master branch have to be updated for this to pass?
Yeah, unfortunatelly they need to be integrated in master first
if you modify the enums in master and ping me as a reviewer, I can aprove the changes
Thanks for the quick reply! So I need to make a branch to edit, then request the PR?
How do you think it would be best to update the wiki on this? To contact community for new enums to be added?
No - anyone can propose additions to the enum, you always just need to make a separate PR
In the 'Pull Request template' there is a sentence that says something like : please post link to enum PR if enum update required
So there is - thank you I will include that now
Could someone please merge “Add files via upload” with the master. I have uploaded a few more screenshots to update the wiki 🙂 thank you!
*Thread Reply:* Done blindly from my phone 😅
*Thread Reply:* HahahBrilliant! Thanks James - wiki is updated and images up to speed 🙂

*Thread Reply:* Thanks for the link, but I'm struggling to log in to ShareLatex... it wants a single sign-on from academiccloud.de?
*Thread Reply:* I have an Overleaf/ShareLatex account already but it's not crossing over
*Thread Reply:* And IT teams wonder why everyone goes to the 'non-data protected' variants of everything
*Thread Reply:* Ignore it then (deleted the message)
Thanks @Ele for updating the step-by-step! - just made a few corrections and updated one screenshot (you caught me out on something Ishould've updated a while agao with the PR template😅 )
And at minimum for @ivelsko and @Åshild (Ash) I'm going to start accepting the open changes this afternoon. So just be aware if they disappear it's not you, just read and compare to the previous version/resposnes if you need
*Thread Reply:* I read through the changes you made and I don’t have anything to add to them or to change
@channel please ignore the links sent to you this morning. They will not work (other than MPI people) 😞
*Thread Reply:* They do for me as I could log in with my old mpi account, but yes, they won’t work for non-mpi people 😐
*Thread Reply:* @Zandra Fagernäs @aidanva you'll appreciate that one 😆
The originals from yesterday should still work, but please make minor-changes to the text directly, and otherwise email/message me with changes
@channel revisions submitted!
@channel FYI I'm planning on doing a new release end of this month. We have lots of open publications to be added, would be really grateful if you guys have a spare 30m if you could assign yourself an issue and make a submission!
I see a few people aleady have assign themselves to a few publications but haven't made PRs yet, and I also have a few open PRs - so if you would rather just do consistency checks also please let me know and I'll assign you to hese!
```Dear Mr Fellows Yates,
Based on the recommendation of the handling Editorial Board member, Lynn Schriml, we are delighted to accept your manuscript entitled "Community-curated and standardised metadata of published ancient metagenomic samples with AncientMetagenomeDir" for publication in Scientific Data. Congratulations and thank you for choosing to publish your work with us.``` 🎉 thank you everyone!!!
Will keep you updated through the proofing steps
@channel please let me know if you have any affiliation changes etc
If you want to see the agreed license
I've requested the OA APC to be paid by the MPG fund, so that will take a few days
And finally, reminder of the open PRs!
https://github.com/SPAAM-community/AncientMetagenomeDir/pulls
And for those who have their assigned papers: https://github.com/SPAAM-community/AncientMetagenomeDir/issues
🚨 The deadline for the next release of the database is the end of this month 🚨
@Ele Did a review on your Morozova PR!
*Thread Reply:* Changes made, thanks for the help!
*Thread Reply:* Will merge in a day or so. A new shiny functionality will be merged and I want to test it on your PR ;)
@channel new functionality: https://github.com/SPAAM-community/AncientMetagenomeDir#current-status
With each new merge to master the images on the link above will automatically be updated. I need to do some tweaking to them (or @Maxime Borry is going to push them to ultra-fancy), but you get the idea.
The images, as with all other content are under CC-BY so you can take them for presentations/grants/papers/reviews etc (juts need to cite!)
*Thread Reply:* Thanks to @Ele for allowing me to mess with her PR to get it working 😄
*Thread Reply:* Cool stuff!
APC is paid! Will send you all proofs when I receive them all!
@Shreya I've reviewed your PR on the Mexico City Treponema, couple of minor changes then we can merge 🙂thank you very much!
*Thread Reply:* thanks James!! done with my teaching for this term so I’m ready to help push this milestone through!
*Thread Reply:* @Shreya feel free to re-request me for review on the PR once you've commited the changes 🙂
*Thread Reply:* Approved and merged, thakn you!
*Thread Reply:* Wooo!!! Thanks James! On to the next!
*Thread Reply:* Both merged! thank you very much!
@Åshild (Ash) @aidanva and @Miriam Bravo you all have self-assigned publications, if you think you won't be able to make the PRs please feel free to un-assign yourselves and we canfind others to do it instead.
*Thread Reply:* I'll make the PR during my day 🙂 .
*Thread Reply:* Don't feel obliged though of course! It's no problem to de-assign yourself.
Hope thesis writing is going well!
*Thread Reply:* Thanks James, is going well, almost done! 🙂
*Thread Reply:* Let me know if you have any problems with the PR btw!
Just sent an email about a request for a blog-post on the paper! Let me know if you would be interested in helping out (if you don't get the email let me know and I'll PM itto you)
*Thread Reply:* I’m interested! I speak English and Spanish
I can help you on this
@channel I asked about the multi-lingual blog-post thing already (saying we would provisionally be interested), and got a positive response:
```Dear James,
I really like the idea of a multi-lingual blogpost, so it would be great if that is possible! I'm happy to look for alternatives if not.
Also, we are implementing policy changes at Scientific Data to ensure more diverse and inclusive datasets as well as increased transparency if data are somewhat biased. I'm always happy to hear your opinion on what we can do at Scientific Data to help the field of palaeogenetics specifically. Also, I 'm more than happy to help promote your Data Descriptor and blog posts when they come out. Feel free to share relevant twitter handles and hashtags with me for promotion on twitter.
Best wishes, Veronique``` So please let me know if you would be willing to at least translate!
I am in for helping and translating
Same, can help translate 🙂
So that's: English, Spanish, Catalan, French - good start!
I'll very happy for translating 😄
I've made a Doodle for the new year to plan about the blog post:
https://doodle.com/poll/vbd3ygyv3s3aypuy?utm_source=poll&utm_medium=link
I should've sent emails to the people have shown interest already, but if you would still like to contribute fill in your avaliability ☝️

@Shreya @Antonio Fernandez-Guerra @Maxime Borry @aidanva @Åshild (Ash) @Miriam Bravo I've selected a doodle date and sent you google calendar invites
for january and the blog post
Thanks @Åshild (Ash) for all the PRs 😱 ! I'll try and review them tomorrow and do a release after they are merged (unless someone else plans to do one by tomorrow?)
@James Fellows Yates Pinar 2020 and Achtman 2020 don’t have anyone assigned to them, but they need to be added, right?
Correct. I think it's just the one from @Miriam Bravo that is open
And Martin2016 bt that's still abit complicated
However we still need reviews on the Ferrari 2020 (thanks for the fix @Åshild (Ash)) and also Lugli 2017 before they can be merged
I’m reviewing Lugli 2017 now
*Thread Reply:* If you're happy, please merge directly
*Thread Reply:* Thank you! ❤️
@Shreya @aidanva @Maxime Borry @Åshild (Ash) @Miriam Bravo @Antonio Fernandez-Guerra reminder we have the call today to talk about the blog post! We can meet here: https://meet.jit.si/LiableExtremesAppealAbsently
https://researchdata.springernature.com/channels/behind-the-paper
https://natureecoevocommunity.nature.com/posts/29300-enteric-fever-in-sixteenth-century-mexico


https://natureecoevocommunity.nature.com/channels/521-behind-the-paper
@Shreya if you PM your gmail I can add you as collaborator 🙂
@Antonio Fernandez-Guerra the plan is I will draft something and share with everyone to lean it up/give feedback. Once we agree to the final thing translators will do the translations
@Shreya @aidanva @Åshild (Ash) @Miriam Bravo @Antonio Fernandez-Guerra @Maxime Borry I've done a first word-vomit of a draft of teh blog post. It's a bit long atm, and I have the feeling the balance is off but you guys can identify the bits to cut down and expand. Please start making comments/rewrites in suggestion mode (and go crazy!)
*Thread Reply:* Is there a link? or will you send it in an email?
*Thread Reply:* Same Google Doc as before
*Thread Reply:* Ok after running it through my head in a dog walk I realised in particularly not happy with the first half. Too rambly, so I'll probably try that again at some point. But you guys can already start working on it already anyway. Maybe someone else does a better job
*Thread Reply:* Don’t worry about it! No one is judging, it’s good that we have something to work with now 🙂
*Thread Reply:* Oh I know. I just meant it might change a lot suddenly 😅
*Thread Reply:* ok cut down at least 100 words from teh beginning. Now I think the last sectoin is too waffly but I'll see what you guys thing
*Thread Reply:* I’ve taken a first pass over the blog post draft, keep what you like, discard what you don’t ;)
@ivelsko and @aidanva / @Åshild (Ash) it would be great if you guys could double check the PR below (randomly pick a few samples, and check the PRJ code is correct against the ERS/SRS code for the hostassociated-metagenome and hostassociated-singlegenome tables respectively (@Becky Cribdon has already checked the environmental one 💪 )
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/350
@channel I'm planning on presenting AncientMetagenomeDir at the Wellcome Trust "Ancient Biomolecules of Plants, Animals, and Microbes" (virtual) conference happening in March 2021.
https://coursesandconferences.wellcomegenomecampus.org/our-events/ancientbiomolecules21/
I have written an abstract with all co-authors of the manuscript as 'co-presenters'. Comments and suggestions are warmly accepted here: https://docs.google.com/document/d/1HvM08gpt5YI8fjfr8XE0dbye35ixYQECb8ulxYh2Uy0/edit?usp=sharing

(Deadline is Feb. 2)
@channel soooo our editor for Scientific Data is really excited for the multi-language blog post (pushing for a solution n how to host it without my prompting 😅 ). If there is anyone else interesting in helping with a translation, please let me know! We have an English first draft almost ready!
Current languages will be: English, Spanish, Catalan, French, Norwegian. (The only two languages I can think from the co-author list of would maybe be@Valentina Zaro for Italian, and @Alex Hübner in German - but again no obligation/pressure!)
*Thread Reply:* I bet our press department would translate it to German, we could ask them
*Thread Reply:* Oh good point
*Thread Reply:* Can keep in mind
*Thread Reply:* I can also translate it into German, no problem.
Oh and maybe Greek by @Maria Spyrou?
It would be something around 750words
Hey James, would love to take part but the deadline is too tight for me..
Hi James, could you remind me when the deadline is?
There isn't one at the moment, ideally around the publication of the paper itself - but it's stuck in a post-holiday production backlog (and we can also publish blog after the publication of the paper)
I would estimate sometime in the next month?
Ok, thanks! I can translate into Italian 😉
*Thread Reply:* Woooho thanks!
OK... proofs are in and we have two days 🤦
I will try and send something around for everyone to see
BlogPost will come after publication I guess, but that's OK!
Any corrections?
@Shreya @Antonio Fernandez-Guerra @Miriam Bravo as we have the proofs already, I guess the blog post should also be made ready soon(ish) [but like I said there isn't an explicit deadline]. If you can at least read through the english version once to leave any general comments I can finalise it for translation 🙂
*Thread Reply:* I’ve made some feedback! Are we still way over the word limit? I can try and copy it into a separate doc and cut it down if that would help
*Thread Reply:* Yes we are, at least based on the original request. If you can make an attempt we can have a look!
*Thread Reply:* You could also just copy and paste the whole thing onto a new page (with a new header)
*Thread Reply:* Instead of a different doc
*Thread Reply:* Ah yes, good call! Let’s see what I can do
*Thread Reply:* Original request was 400-500, but I feel 600-650 might be ok
*Thread Reply:* I’m at 660-- please take a look when you get a chance (and feel free to accept/reject my suggestions as you wish!)
*Thread Reply:* @Shreya I have made a few suggestions to shorten it a bit more
*Thread Reply:* Could you have a look, if so we can adopt that version 🙂, you did a very good job = thank yu!
*Thread Reply:* OK cool, please accept the changes 😄
*Thread Reply:* (if you haven't already)
*Thread Reply:* done! And made one suggestion on where to spell out FAIR
*Thread Reply:* Ok, moved your version to the top
Proof corrections sent for the main manuscript!
@aidanva @Åshild (Ash) @Antonio Fernandez-Guerra @Miriam Bravo @Maxime Borry Shreya did a good pruning pass and cut down the blog post to ~650 words, which is better in range of the original request. We've entitled this 'short version' in the Google Doc, please read through it and see if you're happy with it. If you are, we can start doing the translations! I think we will be published-published relatively soon!
The communities people replied: we will put all the translations on one page, and somehow indicate at the top the different languages.
So once I have OKs from @aidanva @Åshild (Ash) @Miriam Bravo and @Maxime Borry on Shreya's short version we can begin translation and I'll start preparing the blog post on the website interfact
@Shreya’s short blog post version looks great to me! No further comments
One note about ORCIDs (as originally asked by @Åshild (Ash))
"It has been noted that you have requested to add ORCID information to a number of your authors on your returned proof. It is Nature Research policy to only include ORCID information in the published article if it has been added to the author’s profile on our manuscript tracking system prior to formal acceptance. This is to ensure the accuracy of the ORCID information: before acceptance, each author wanting to add their ORCID is asked to log into their ORCID account from our manuscript tracking system; the ORCID information is then stored and passed on to our production system. This process validates the veracity of the ORCID information. ORCID IDs added outside of the manuscript tracking system, e.g. by email or at proof, will miss this important step of validation.
Please note that although these ORCIDs cannot be displayed on the published paper, authors are able to add the published work to their own ORCID records. For further information please see the ORCID help website (https://orcid.org/help)."
So if you want the paper associated with your profile, you have to manually add the paper to the profile after the paper is out. Also it's good practise whenever you sign up for any form of academic account always check for ORCIDs as one of the first things you do, so you don't miss it. You'll be surprised where you can miss it, but it can add a lot to your profile and CV! e.g. Zenodo objects can be associated there too (and you can get download stats of the resources you upload for your CV)!
Also @aidanva @Åshild (Ash) @Miriam Bravo @Maxime Borry @Shreya @Alex Hübner @Valentina Zaro I've made a new document and shared to your respective gmails for blog post translation (Valentina, please PM a gmail/google account if you have one, and I can share with as a direct collaborator, if you don't have one I'll send you a link - let me know either way)
*Thread Reply:* thanks James! I’ve written to the Spanish team so we can make a plan!
*Thread Reply:* Organisation 💪💪💪 I love to see it!!!!
@James Fellows Yates I won’t have time in the next few weeks to get this done I’m afraid. Can we add translations later on?
*Thread Reply:* We can't edit the blog post I think. But maybe Norwegian isn't so crucial for impact 😅
*Thread Reply:* But we can see how long the other translations are taking
*Thread Reply:* Yes, I think the five Norwegians who do aDNA all speak excellent english, so impact will be low.
*Thread Reply:* I might give it a miss if that’s ok
*Thread Reply:* Also, haven’t written in Norwegian much since I was 14, so it would take me a whole day to get this done, which I just don’t have atm 😅
@James Fellows Yates - The french translation: https://docs.google.com/document/d/17jfznirdkzG_WV7UkAkE9Eql752TW3W3SedwPO8ZPz4/edit?usp=sharing
I'll transfer that over to the main documentation
@channel Publication date of the paper - 26th Jan!
Want to check with the Press-Departement @James Fellows Yates?
*Thread Reply:* I'm honestly too tired atm 😆, if you would be willing to do so that would be great! I'm also not sure if ti's something that would be interesting for them 🤔
Reminder for the blogpost team: https://docs.google.com/document/d/1YxmFZ3IuQHZInuKLnKMzTQco34NOYwZL6a9_40VAoFA/edit?usp=sharing you can insert your translations in that document! (thanks already to @Maxime Borry and @Alex Hübner)
I've invited all the blog-post team to the actual blog post on the Springer Nature communities website. That way you can update your profile with your face etc. for the author list
Ok, good news, I can do in-post links, bad news, you can't save draft posts -.-
therefore the link I sent hopefully should still allow you to make an account, but be aware the blog post itself (if the registration emailt includes it), won't work
I'm going to add the Italian translation this morning 😉
We (the Spanish team) have finished the translation, I just added it to the above document.
*Thread Reply:* Woo thank you very much @Shreya and @aidanva as well!
https://www.nature.com/articles/s41597-021-00816-y
Congratulations everyone!
If anyone has time to do some Twitter PR I would be grateful,!
(Maia is insisting I'm a baby and that I have to sleep)
Ok they did it for us: https://twitter.com/ScientificData/status/1354059031883747330?s=19
All translations are in, one needs a proof read. Current plan is to post it one week from today
FYI I've submitted to the abstract to the Ancient Biomolecules 2021 conference!
@Antonio Fernandez-Guerra @aidanva @Åshild (Ash) @Miriam Bravo @Maxime Borry @Shreya @Valentina Zaro @Alex Hübner REMINDER Please register on https://researchdata.springernature.com/ so I Can add you to a contributor to the blog post!!!!! It will go up next Tuesday!
*Thread Reply:* Do I need to do anything after I registered ?
*Thread Reply:* not for the itme bein
*Thread Reply:* I should be able to add you
*Thread Reply:* Let me know if it comes up in your posts thing
*Thread Reply:* Registered there as well
*Thread Reply:* Haven't seen my posts anywhere
*Thread Reply:* Click on your face > My Posts
*Thread Reply:* Nothing listed?
*Thread Reply:* I’ve registered :)
*Thread Reply:* I've registered too!
*Thread Reply:* Done! (with my gmail: shreyarama42@gmail.com)
*Thread Reply:* Found you! Just @aidanva and @Miriam Bravo to go and we will be pretty much ready 😄
*Thread Reply:* I registered with twarinner@gmail.com
*Thread Reply:* I registered too (bravolomiriam@gmail.com)
*Thread Reply:* After your pestering @James Fellows Yates, I also registered
Otherwise I'll just list you with no linkin function in the header
Thanks for the much more interesting tweets that my ones @Åshild (Ash)!!!! https://twitter.com/AshAshild/status/1355200941448974338?s=19 👈for anyone else who wants to support the word
@Åshild (Ash) we should add your last tweet as a quick start guide to the main readme! Could you do that?
If it make it on a branch I'll expand it for getting days from Zenodo(rather than the latest)
OK @channel Blog post is now scheduled for going live at 11:30am GMT Tomorrow! I will try to tweet about it from the shitty phone connection from my new apartment. Otherwise keep an eye out for it and fire away!!
*Thread Reply:* “Unfortunately you are not authorized to view that page” but perhaps I will be tomorrow!!! Excited for the big multilingual reveal!!
*Thread Reply:* Yeah, I couldn't find a way to add anyone else as 'editors' unfortunately
*Thread Reply:* gotcha! Well I will look out for it!
Ok, i completely failed to catch up on it. I think @Maxime Borry already did (thank you!), I'll also tweet about it tomorrow!
And thanks @Shreya!!
I'll schedule a tweet for tomorrow too
@Maxime Borry @aidanva @Miriam Bravo @Shreya @Alex Hübner @Valentina Zaro we should send language-versioned tweets of the blog-post as well! Feel free to send here the link to tweets you make (so we can all rewteet) or translations if you don't have twitter
*Thread Reply:* Thanks @Miriam Bravo!
*Thread Reply:* I don't have twitter but I can send here the translation if anyone wants to tweet in Italian! 😁
*Thread Reply:* @Valentina Zaro if you send it to me here I can tweet it tomorrow!
*Thread Reply:* Sei curioso di scoprire come in soli due mesi (e nel mezzo di una pandemia!) è stato possibile passare dall'idea alla pubblicazione di un database di riferimento per il DNA antico? Dai un'occhiata al breve "dietro le quinte" della nostra esperienza con AncientMetagenomeDir!
Just read the blog post - it’s brilliant! Well done everyone involved 👏👏👏
@Valentina Zaro you need to make it 19 characters shorter 😅
Oh no wait, I have an idea!
Yes, I'll quote retweet my English one and use your text in the quote!
Sorry, I didn't know about the characters 😅
No problem! Have the solution :), will post tomorrow around 11
https://twitter.com/jfy133/status/1357276243310350336
(@Valentina Zaro, I also posted your researchgate profile to saw who wrote the tweet, I hope that's ok)
(you can see it if you click on the link)
Just saw the tweet! 😁
@channel I'm in the middle of moving to a new apartment at the moment and will be having to play a lot of catch-up of other stuff next month. Would anyone be interested as acting as coordinator for the next release (should be sometime in march). This would involve ensuring as many open issues get added to the database (both doing one or two yourself, and distribute to others), and then doing the release itself (I've written documentation for this as well, and would be available here if something is unclear). With the release you can leaen a bit more about how github works and how it integrates with Zenodo to auto archive stuff.
I will be presenting AncientMetagenomeDir as poster and lightning talk at the WellcomeTrust thing.
"Please submit your poster (1 page) in a PDF format. The max file size we can accept is 8mb. There are no other specifications in terms of size or orientation for posters."
Tempted to do a circular one 😂
*Thread Reply:* That... could actually be fantastic.
*Thread Reply:* I shouldn't... buttttt
*Thread Reply:* Why not? It would be eye-catching, and would lend itself to presenting several connected concepts around a central idea. That would work well for an introduction to something.
*Thread Reply:* FINE! I'll try and inevitablly waste my evenings on it 😉
*Thread Reply:* But I ain't the one making the poster so 😁
*Thread Reply:* Are you familiar with this person? Probably to blame for my liking of simple but outlandish posters: https://twitter.com/Better_Posters
*Thread Reply:* That person who does the big title/core result in big bold right in the middle?
*Thread Reply:* Ah, that's Mike Morrison. Better Posters did a critique of the Morrison poster but also consider other approaches: http://betterposters.blogspot.com/2019/04/critique-morrison-billboard-poster.html

@channel we have 2 more issues open and we can do a March release, if anyone is up for it?
https://github.com/SPAAM-community/AncientMetagenomeDir/issues/365 https://github.com/SPAAM-community/AncientMetagenomeDir/issues/364
(I'm also back to reality, but if anyone is interested in learning about the release procedure, please ping me!)
I've made a first(!!!) draft of the poster for the Ancient Biomolecules conference thingy.
Thoughts/feedback welcome!
(Couldn't get a circular design to work unfortunately, but decided to try out an 'infographic' style instead for funsies. Also: before the perfectionists arrive, I know things are off-set or not aligned ;), that will be fixed later!)
Looks very nice although there are things off-set or not aligned 😁
Looks great to me too, although I have a few comments: 1. In the tsv file screen shot appear the the wavy red lines as they were grammar errors, maybe you could replace that? 2. In the What is in it? section, it will be more clear to add the sample number for microbiomes, microbial genomes and sedimentary genomes.
*Thread Reply:* Thank you!
*Thread Reply:* You're welcome. By the way, which software did you use to make your poster?
*Thread Reply:* https://inkscape.org/
*Thread Reply:* Icons from openmoji.org
*Thread Reply:* Or drawn by @Antonio Fernandez-Guerra
*Thread Reply:* @Antonio Fernandez-Guerra where can I find more info about drawn?
*Thread Reply:* The icons we used for the workflow are from here https://primer.style
*Thread Reply:* https://octicons-primer.vercel.app/octicons/
If anyone has a spare 3 seconds to approve this: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/371
*Thread Reply:* Ok Aida got there already 😛 thanks though!
I need to it before adding the screenshot to the spoer
Ok new version does this help?
(in terms of alignment, and spacing issues?
Ok, some people have said the TSV table screenshot is too small and is maybe not necessary.... any suggestions what to replace it with?
Looks great, @James Fellows Yates! re. TSV table: if you move the ‘sample age’ column to the lower panel and drop the lower two samples, you should be able to make it both larger and with the upper and lower panels balanced.
*Thread Reply:* Is the question mark at the last point of the 'needs improving' section necessary?
*Thread Reply:* Yeah because it raises it the quesiton if other fields would be interested in that or not...
*Thread Reply:* We would do it ourselves (hopefully)
*Thread Reply:* but question is if we should already involved other firleds
If there are no more objections I'll submit it this afternoon 🙂
*Thread Reply:* Lag:Long @James Fellows Yates
*Thread Reply:* (instead of Lat:Lons)
Last PR for today's release ('cause I totally forgot)!
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/378
Release v21.03: https://github.com/SPAAM-community/AncientMetagenomeDir/releases/tag/v21.03
@channel So as a part of PhD theses as FSU Jena, we apparently have to specify 'how much' we contributed to various aspects of each paper.
I have no idea how one is expected to quantify most of these columns, but I've made up some semi-random BS to make this work - if any of you have an issue with any of the assigned values in the table below, please PM and I can modify accordingly. The only one that has any resemblance to reality is the Experiment (i.e. generating the data) which I partially based on GitHub stats (but then tweaked so I can round up to whole numbers). The rest I have no clue e.g. I have no idea how much each person contributed to the paper writing)
Just wanted to give everyone the opportunity to have their say and that it doesn't appear in their google scholar alerts in a few months time with no warning, and I upset anyone by them thinking I undervalued their contribution!
If anyone has a spare 20 minutes:
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/386 https://github.com/SPAAM-community/AncientMetagenomeDir/pull/385
@channel I've now worked out how to partially automate extracting library level metadata from the ENA/SRA. We now need to work out exactly what metadata would be useful in such a sheet, which would also include all the FTP links to each library.
I've made a github issue here, please see my suggestions, and then leave comments what else we should also add, and we can start making column specifications 👍
https://github.com/SPAAM-community/AncientMetagenomeDir/issues/388
My suggestion(s)
Required:
- [ ] Sample name
- [ ] Library name
- [ ] Shotgun vs whole-genome-enrichment (latter for pathogen/single host genome ONLY)
- [ ] Library type (SS/DS)
- [ ] Library treatment (e.g. UDG)
- [ ] Sequencer type
- [ ] Pairment (single- or end)
- [ ] Cycles (i.e. read length, 50, 75, 100, 150, 300 etc)
- [ ] ENA code
- [ ] Type of raw reads (all shotgun, mapped only)
- [ ] FTP link
Possible
- [ ] Lab
- [ ] Extraction and Library protocol
- [ ] Library concentration or qPCR values? (useful for microbiome contamination estimation e.g. in [decontam](https://github.com/benjjneb/decontam))
Another thing I thought of was read count
Any opinions @channel ☝️ ? What would be useful information for you to have about libraries, when adding previously published samples to your own analysis?
Ok! @channel (sorry to bother you again) - it's June which means it time for a new release of the 'Dir.
We still have quite a few issues open (and one open PR). Unfortunately I don't have time to do them all, so I would be really grateful for help this time 🙏 . In particular there are a lot of environmental related ones (@Pete Heintzman, @Vilma Perez, @Anneke ter Schure, @Becky Cribdon, @Linda Armbrecht for example), and a couple of outstanding pathogen ones (I see Ash and Aida have assigned themslves to a couple but there is still one open - @Miriam Bravo, @Maria Spyrou, @Shreya, @Jessica Hider maybe?). So I'm hoping there are volunteers for those? Only need to do one each!
*Thread Reply:* on it! will probably only get to it when the europe squad is asleep though!
*Thread Reply:* No problem !
*Thread Reply:* I can help out! I'll just have to remind myself how to do everything because I've been naughty and not checked in recently (good thing there are awesome tutorials 😉).
And another reminder for input the discussion about which library level metadata you would like for automatic downloading of files (see link above)! @Marcel Keller I tink you had some opinions on this?
OK I think I missed a trick there 😆 (@Maria Spyrou you should name stuff in the future :D)
No no I love it! that's what I'm going to call it in the future 😆
my notorious typos .. 🙂
I love the idea of a Megagenome!
Thanks @Pete Heintzman! Gonna get to them this evening ❤️
@aidanva @Shreya @Åshild (Ash) @Miriam Bravo reminder that you assigned yourself issues. I would ideally like your PRs in the next week (if possible) to give a week for mistake corrections before the end of the month 🙏
First draft of automated library metadata construction!!!!
Thanks @Shreya will look this evening or at the weekend
thanks James! also a shoutout to @Ele for the guide which continues to be exceedingly useful
Random weekend fun fact: ancient host associated metagenome libraries are typically sequenced to 4.8 million reads.
@aidanva @Åshild (Ash) @Miriam Bravo reminder of end of this week deadline so we can make the june release end of this month!
Also, you guys can also help: https://link.springer.com/article/10.1007/s12520-021-01350-z Y/N for inclusion? can't really tell if it's sufficient...

*Thread Reply:* I will work on my PR today. About the paper, I would not include it, they don't have enough reads and they even say that they can not authenticate the aDNA... but that's just my opinion
*Thread Reply:* Ok we leave it off then 🙂 thanks!
*Thread Reply:* Given that this is the coverage they have across the mtDNA after capture, I definitely think we should not include it (from their supplementary).
*Thread Reply:* Tha'ts what I thogught 😄
*Thread Reply:* I'll do my PR today 🙂
Hi folks! Apologies for not checking this lately - my PhD is down to thesis corrections (thank you @Pete Heintzman 👍 ), I'm up to my ears in other work, and I'm shortly moving out of academia. I'm afraid this is my resignation from SPAAM and the 'Dir.
It's been an absolute pleasure 🙂
*Thread Reply:* No problem - good luck with the corrections and your future endeavours! Thanks for all your contributions and support ❤️
*Thread Reply:* Best of luck for the future, @Becky Cribdon!
@channel I've just realised collection_date might be misleading, as this refers to the explicit sampling of the sample, not when the sample was taken from the ground or collected from the wild. Any objections to changing it to sampling_date?
The purpose of this column is to make sure people know when it was previously sampled, to prevent repeated sampling for the same purpose
*Thread Reply:* I think it's good to change it
*Thread Reply:* OK, that's three yes's - gonna change it now!
*Thread Reply:* Requested you all as reviews ❤️
*Thread Reply:* accepted and merged
*Thread Reply:* Btw, Zavala is no go
*Thread Reply:* " the raw data for each mammalian mtDNA and human mtDNA enriched library are available in the European Nucleotide Archive under accession number PRJEB44036. "
Also can anyone quickly approve tihs: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/410
*Thread Reply:* done and merged!
*Thread Reply:* @aidanva one more: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/411/files
*Thread Reply:* Forgot the host species 🤦
This would be for ALL tables
If anyone ha spare thirty seconds: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/413/files
This is to make a paritcular git log file MUCH smaller (currently it makes the repository 200MB for 4 TSV tables... 🤦)
Is the Piñar 2020 paper problematic? or has it just not been done yet?
Just not done. Originally there was an undergrad student who wanted to help out and we had tasked them to do the anthropogenic stuff but then nix. Given there isn't much movement in that research area (as far as I can tell), I've not focused on it for the time being.
https://twitter.com/jfy133/status/1409485777172676611
Hi All,
this was sent to MPI-SHH yesterday:
https://www.stifterverband.org/innosci/open-data-impact-award/english
, and a couple of us here thought that while it might be a long-shot (as the benefits are primarily academic), that we could maybe put forward AncientMetagenomeDir for it.
If it got through we could use money to boost SPAAM projects/workshops etc. So if any of the more active contributors are interested in contributing to writing the application please let me know and I'll send you a doodle. We would have a meeting to plan/write last week of July.
*Thread Reply:* I think it’s worth a shot! It’s only a (max) two page application. Happy to help out!
*Thread Reply:* Will send you doodle invite!
*Thread Reply:* Let me know if you got it @Åshild (Ash), send to your gmail
Thanks for the PR @Jessica Hider! Reminder: you can modify multiple files on a branch (you don't need to make a separate one to update like the changelog, if you forget the first time around. You can modify always PRs in situ)
And still waiting for a review on this minor typo correction: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/416/files
*Thread Reply:* Thanks @ivelsko
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC8278039/
y/n?

@channel Aida, Irina, Maxime and I have drafted our application to the open data award thing. We would welcome your comments/feedback (it's only 2.5 pages) as we would need to submit this at the *end of this week*.
Feedback from anyone with experience with grant writing (e.g. @Christina Warinner) would be particularly useful as us 4 have not done that, but comments from everyone will help a lot! @Hannes I've made a reference to maybe helping put the data into AncientGenomes.com, if you're happy with us saying that?
*Important* we are over the limit by 0.5 pages, so places to cut superfluous stuff would be welcome!
https://docs.google.com/document/d/1edxlM4dxdX2vcuCWhFiNyxu55lfubSjRHVoOfTl6mT4/edit?usp=sharing
Great job all! It reads very well
Application submitted, final version is here if anyone is interested: https://drive.google.com/drive/folders/1F_kHelCjAI_hPT90TSYOnvjV-kY9reaN?usp=sharing
Thanks to everyone who contributed!
@channel who is attending SPAAM3? I need input on the next stage of AncientMetagenomedir, so was wondering if could arrange a brief/informal chat about it.
I have ready the majority of the raw library metadata information, and will need help in cleaning it up and filling in the gaps.
Importantly, this metadata will include links to FASTQ files so we can build tools to automatically download datasets for you 😉
*Thread Reply:* Is this the attempt to download the sample/seq. run information from ENA/NCBI that contains all the information on each uploaded library (read count, library type, FastQ/Aspera download URLs)? If yes, I would be interested in.
*Thread Reply:* Yup, but even better: in addition polyermase/UDG treatment etc. which can tell you about levels of damaage etc.
*Thread Reply:* Hi @James Fellows Yates. Sounds great and I'd also be interested though unfortunately won't make spaam3 today due to attendance at another conference.
*Thread Reply:* Ok, I don't know yet when we will meet but can let you know when it happens and you could join if you're free.
I can also summarise what we discuss in the end.
The main thing for me would be finding out what would be good incentives for people to contribute time in cleaning up the data
@channel for more clarification - we currently have 117 papers in AncientMetagenomeDir, so if for example I had at least 10 volunteers, we could each take 10 papers to clean up the tables I've already pulled from the ENA, which would massively reduce the workload. I think with 10 papers per person, we could probably do it all in one day.
But of course I understand most people won't want to invest time into something that may not directly go towards your work.
So I would like to have as uch input as possible from you guys on what would be good incentives for you to contribute. (Pizza? Stickers? ECTS points? A publication [although this would be more difficult/long term, as we would need to find a way to differentiate from the first paper])
Note: volunteers can be people who weren't in the original release!
*Thread Reply:* I'll try to get through some next week!
*Thread Reply:* Oh it's not ready yet 😅
*Thread Reply:* the new structure/raw data is on my fork at the moment
*Thread Reply:* But once we have the volunteers we can arrange integrating the new version into the main repository 🙂
*Thread Reply:* I mean I love a good sticker but I’m also just happy to power through 10 papers
*Thread Reply:* I work for stickers....and pizza....no stickers...no pizza....STICKERS!
*Thread Reply:* idea: what if editors hopped on zoom one day and did a table-a-thon with a couple breaks for games
*Thread Reply:* I was thinking EXACTLY along the same lines 😉
*Thread Reply:* Is that something that your supervisors/PI would agree to?
*Thread Reply:* Mine at least is very supportive of my SPAAMy activities! And I doubt it’d need more than 1 day of working hours
So if you aren't attending SPAAM, please also just respond here 🙂
So currently including me we have 8ish volunteers including me. Anyone else?
*Thread Reply:* You're already included @aidanva 😉
*Thread Reply:* I would be interested but I don’t know yet when I’m available.
*Thread Reply:* Just fill in the doodle, you should have optin of 'maybe'
*Thread Reply:* done 🤡
@Adrian Forsythe has joined the channel
@channel for those who are new to AncientMetagenomeDir and want to get familiarity already with the already-established structure for samples etc. before we do the library metadata metadatathon, we still have a few publications that have not yet been added to the sample metadata tables!
If you want to practise before the metadata-thon
1) please register with github (if yuo've not already), and send me user name so I can add you to the github organisation.
2) Then you can check the various publications in the issues page, and assign yourself for to one of the open publications 👉 https://github.com/SPAAM-community/AncientMetagenomeDir/issues
3) Follow the lovely step-by-step guide from @Ele to add a publication 👉 https://github.com/SPAAM-community/AncientMetagenomeDir/wiki/Adding-a-Publication:-Step-by-Step-Guide
*Thread Reply:* Should have an invite 🙂
*Thread Reply:* Or go: https://github.com/SPAAM-community
*Thread Reply:* Pretty sure you type quicker than I am able to think 😆
*Thread Reply:* I'm not even that fast 😱
*Thread Reply:* You should have an invite :)
@Gunnar Neumann has joined the channel
@Raphael Eisenhofer has joined the channel
@Nikolay Oskolkov has joined the channel
@channel we've got the data for the metadata thon!
Please mark your calendars as NOVEMBER 5th 2021
I will start planning the introductory sessions, and maybe for those in Pacific timezones I can do that the afternoon/evening of the day before; then intro session morning for european-based peepz, and then an morning for the Americas
For those using google calender: https://calendar.google.com/event?action=TEMPLATE&tmeid=M2M3Z2UwazRzcXY1am1mbXJrZXBrOW80azggamZ5MTMzQG0&tmsrc=jfy133%40gmail.com
(if the link doesn't work, send me your gmail account and I send you an invite)
I can´t open the link here my email bbmoguel@gmail.com thank you.
@channel reminder, we are scheduled for another release end of this month (next week!)
If you want to practise adding sample metadata (Before the library metadatathon in November), please assign yourself to an issue here:
https://github.com/SPAAM-community/AncientMetagenomeDir/issues
A walkthrough guide of the whole procedure is here: https://github.com/SPAAM-community/AncientMetagenomeDir/wiki/Adding-a-Publication:-Step-by-Step-Guide
I just added another paper, so we still have 4 open papers to be added (all from bacterial genomes!)
⏰ deadline for this months release is drawing near!
I had a quick look over the weekend and realised most of the remaining publications were like one sample each, so I made PRs for each of them, so there are *6* open PRs waiting for review!
I would be very grateful if anyone could look through any them and check they look OK <3
*Thread Reply:* Hey James, happy to help out if you could tell me what to do! 🙂
*Thread Reply:* Wonderful! I'm just finishing a tutorial on how to review, and will post it to you
(4 pathogen papers, 1 microbiome, and 1 sedaDNA)
If you're new to hte project and want some guidance on what I mean by review, please let me know and I'll get you up to speed!
Thanks @Pete Heintzman @Barbara @irinavelsko
Just four more to go and we can release 21.09 'Taputapuātea' 🙂
@Barbara don't forget to accept the GitHub inviitation! @Meriam van Os I've just sent you a GitHub invitiation, let me know i you didn't get it
*Thread Reply:* ohh I thin my invitation expired. :(
*Thread Reply:* Oops I will try again
*Thread Reply:* @Barbara try again now!
*Thread Reply:* I resent it
*Thread Reply:* I am in!! 🙂 thank you!
*Thread Reply:* I will check the tutorial. I am hurry with an inform but finish that I will be more active in the analysis! I really want to lear!
OK! I did an @Ele written a tutorial on how to review AncientMetagenomeDir PRs 😄!
https://github.com/SPAAM-community/AncientMetagenomeDir/wiki/Reviewing-a-Pull-Request:-Step-by-Step
Please try it out and review some of the opens PRs, also send me feedbcak if there is anything that is not clear/could be reprheased/ascreenshot is missing
I'm NOT good at writing, so any improvements welcome 😉
Ping in particular @Meriam van Os @Sina White @Bjorn Bartholdy who should interest in helping out 🙂
(and maybe @Anan Ibrahim @Jasmin Frangenberg @Olivia @Gunnar Neumann @s.wasef @Ophélie Lebrasseur @Megan Michel @Ian Light @Adrian Forsythe who all signed up for the library metadatathon <- note the current reviews are for sample level, but the same principle will be used for the library metadata during the day, so if you want to practice now is a god time! )
Thank you very much @Meriam van Os for the reviews, they are perfect 👌 (I've made the changes and rerequested the review :))
Also: uncorreted proof is fine, that means it's accepted 🙂
If there is anyone else who wants to try a review before the end of the week, please assign yourself to the PR (on teh righthand side of the PR)!
No worries @James Fellows Yates, let me know if you want me to have a look at the others as well (I thought I'd leave those for now so someone else can have a try). Walkthrough was good yep! I found a few wee spelling mistakes, but nothing major, and everything was clear to me! 😊
*Thread Reply:* Did you fix the spelling mistakes already? You should be able to aedit the page
*Thread Reply:* No haven't, but can do next week though!
OK thanks! So we can leave the other PRs to people like @Bjorn Bartholdy;).
(@Bjorn Bartholdy if you wlil have time today, can you do both, and then I can do the release today - just one day late 😉)
*Thread Reply:* Spent my morning converting unix commands to harry potter spells... I suppose I could do something more productive... 😅
@Meriam van Os Wu 2021 is now ready too :)
*Thread Reply:* There was just one last thing with one of the dates, sorry... Have a look and see what you think, otherwise all done!
*Thread Reply:* @Meriam van Os I don't see any comments about dates I hadn't resolved?
*Thread Reply:* Ah it came up now. Will look this evening 👍
*Thread Reply:* Sorry, I forgot to klick on submit review...
*Thread Reply:* No worries 🙂 done it myself too,
*Thread Reply:* To respond: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/439#discussion_r720768505
('ve resolved but without change)
*Thread Reply:* But actually, while that one was OK, XBQM11 so I'll update that now (I think I copy and pasted the wrong value 🤦 )
*Thread Reply:* And the NAs are inferred archaeologically from the rest of the text
*Thread Reply:* REquested re-review, and maybe I can do a release tomorrow if all good now 🙂. Thanks for your very careful checks @Meriam van Os!
*Thread Reply:* Woohoo thank you!
Thanks @Bjorn Bartholdy! So just Meriam to do the final approval of Wu and I'll release - thanks very much everyone!
@channel I'm starting to plan the metadata-thon.
For those who will be joining on Friday. November 5th, I would ask you to kindly answer some questions on the following google form for organisational purposes (timezones etc.):
if you didn't previously respond, but would since like to join and contribute: please feel free to fill in the form! More the merrier! We will be cleaning up library-level metadata and making a tool to assist in downloading the data :).
https://forms.gle/wupVap56sHuDS28q7
Aaaaand AncientMetagenomeDir release v21.09: Taputapuātea is now live!
Release v21.09 includes 9 new publications, representing 122 new ancient host-associated metagenome samples, 13 new ancient microbial genomes, and 12 new ancient environmental samples.
This brings the repository to a total of 132 publications, 1038 new ancient host-associated metagenome samples, 368 ancient microbial genomes, and 375 new ancient environmental samples.
And due to some GitHub magic, contributors are now listed in the given release :)
https://github.com/SPAAM-community/AncientMetagenomeDir/releases
If anyone wants to practise submitting data, here is another one: https://github.com/SPAAM-community/AncientMetagenomeDir/issues/444
@channel The Metadata-A-Thon day is two weeks away! I aim to send more information this friday. But in the meantime I've since added two more publications that can be added at *sample* level, so please use these to practise if you have not already!
In particular @s.wasef @Marcel Keller @Ophélie Lebrasseur @Gunnar Neumann @Olivia @Megan Michel @Adrian Forsythe @Raphael Eisenhofer (who all inidicated they hadn't submitted stuff before). For those working on environmental DNA feel free to post an issue with a paper that is not yet included as well
@channel you have one hour to vote what type of place you want to work in for the Metadatathon 😬 ☝️
Ok, closing with a forest office!
*Thread Reply:* Almost as inspiring as medieval tavern 👻
More details: https://spaam-community.github.io/#/events/metadatathon-nov2021/README
By more details I mean, schedule, location etc!
Now listed on bio.tools 😎 https://bio.tools/ancientmetagenomedir
Also slides for Next Friday: https://hackmd.io/@jfy133/HyKgEJVLY#/
If anyone wants to look through and let me know of anything unclear/confusing and I will update them before the event 🙂
Update on the open data award:
We were shortlisted but didn't make it. But encouraged to apply again next year (I suspect they want to see some more real world use)
*Thread Reply:* A shortlisting is a win!! Yay for us!
If you want to practise a sample-level PR before Friday, here are the open Sample-level issues:
• https://github.com/SPAAM-community/AncientMetagenomeDir/issues/440 (this has been added at metagenome level, but possibly needs single-genome) • https://github.com/SPAAM-community/AncientMetagenomeDir/issues/445 • https://github.com/SPAAM-community/AncientMetagenomeDir/issues/447
Cross posting the email:
```Dear all,
(if you've already let me know you can't make it, please ignore, just reusing old email list 😬)
Thanks again for signing up for the AncientMetagenomeDir (horribly named) metadatathon happening THIS FRIDAY (NOV. 5th)
I think I have pretty much everything ready now, with the details here: https://spaam-community.github.io/#/events/metadatathon-nov2021/README
In particular please check the Prerequisites section to make sure you have everything ready.
We will be meeting on gather.town here: https://gather.town/app/biUhnBjsbwZexQ8w/ancientmetagenomedir
I will do three intro and training sessions to cover each time zone:
On November 4th 09:00 CET (Nov 4th 18:00 AUS) I will do an intro/training session for those who are in Australia. On November 5th 09:00 CET: We will do a handover from AUS, and I will do an intro/training session for the European (+MEA) timezones On November 5th 14:00 CET: I will do an intro/training for the Americas
After each training session is when we do the actual metadata cleanup/retrieval :).
Please let me know if you have any more details!```
Regarding PR reviews: When approving a PR with only minor/cosmetic changes, the branch will get merged without those changes, right? Or is the idea to contact the submitter also to fix those?
So you can approve with minor/cosmetic changes, and ask the person to correct them before they merge it in
Corrected the email above 🤦
For Friday, if you're new to git this Twitter thread is a good start to get familiar with some of the different terms we will be using!
@channel for those in Asia/Pacific timezone or want a headstart we are starting the welcome/training in a couple of minutes!
https://gather.town/app/biUhnBjsbwZexQ8w/ancientmetagenomedir
Ping primarily @Raphael Eisenhofer 😬 as the last person who I think is in AUS?
@s.wasef btw I reviewed and merged your PR already. I found a lot of bugs in my original back end stuff which I ended up fixing on your branch 😅, so I didn't want you to go through everything I fixed.
You can start the next publication when you start again :)
Oh and in addition I completely forgot I wrote a whole tutorial with tips&tricks for adding library stuff (it's slightly different as the _RAW table isn't used here, as I will be automating that being given to you when someone add ssample metadata), but the genreal process is the same. You're also welcome to update/modify that if anyhting isn't clear!
Good morning!
17 PRs 😍😍😍 thank you very much @Raphael Eisenhofer @s.wasef @Meriam van Os!
I've just done a couple of approvals for the sequencing centres from my phone, but you'll need to resolve the merge conflicts before merging it in (make sure to keep both lines that are conflicting), then you can re-run the tests :)
Ill aim to get there for 8:30 CET and we can chat a bit before the Europe shift starts :)
If you don't have permission to merge into master one the conflicts are resolved, let me know and I can do that from my phone (will be a bit sporadicc while I get child/dog ready for the day)
*Thread Reply:* Hey, looks like I can't merge it
*Thread Reply:* Which one did you resolve?
*Thread Reply:* We will have to go one by one through the 3 sequencing centres PRs
I won't be able to make it to the handover at 9:00 CET (doing groceries/having dinner 🍄)
*Thread Reply:* No worries! Thanks for your contributions!!
*Thread Reply:* Thanks for coordinating this fantastic effort! I'll do a bit more tomorrow
@s.wasef @Raphael Eisenhofer @Meriam van Os it's pissing down with rain here and my dog has decided fuck that, so I'll be on gather town in 15m if you wanna talk about anything
*Thread Reply:* No worries at all !
*Thread Reply:* Only come when you have time!
*Thread Reply:* I reexcuted the tests for Schulte btw 🙂
*Thread Reply:* So yu can labbel that for review hwen you have time
Btw People who are in the same institute, feel free to meet/sit together in real life (😱 ) as well as on gather.town as you wish. (EVA people: I'll be staying at home)
For the people at MPI-EVA: I am sitting in the Terrarium if you wanna join me 😄
@Raphael Eisenhofer I'm fixing in all your PRs a booboo I made in dev 🤦
So don't worry if yo usee commits fro mme
@channel We should be starting in 1 minute: https://gather.town/app/biUhnBjsbwZexQ8w/ancientmetagenomedir
Would anyone be willing to explain the difference between sample vs library to a beginner?
*Thread Reply:* I will try: your sample will be the element that you produce your extract from
*Thread Reply:* from that extract you can produce multiple libraries
*Thread Reply:* if it is not clear, come find me in gathertown 🙂, I am by the raised beds
*Thread Reply:* Yeah, sample is e.g. bone or tooth, Library is the DNA from the bone/tooth with sample-specific ID tags on it
Hi @channel important enzyme announcement for the library metadata entry!
For the column library_polymerase you should enter the polymerase that was used for the amplification while adding indexes and not for the adapter attachment
(so not T4 polymerase/Bst, but rather PFUTurbo/KappaHiFi/etc)
*Thread Reply:* so we put the whole “Pfu Turbo Cx Hotstart DNA Polymerase” or I assume rather “Pfu Turbo Cx Hotstart”?
*Thread Reply:* Yep, the 2nd one is good
*Thread Reply:* and does anyone know the one they use for ss-libraries in Leipzig? so i save time looking it up
*Thread Reply:* “Owing to the low concentration of template DNA in these indexing PCR reactions, a hot-start enzyme, AccuPrime Pfx DNA polymerase, is used to suppress the formation of primer dimers.”
*Thread Reply:* from the Gansauge paper
Sorry about that. I'm updating the README/JSON specificaications and already merged libraries now. This was a misunderstanding on my part
what if library-ID on ENA does not correspond to the pandora ID? i guess i leave the ENA id because others wont know anyways..?
*Thread Reply:* Can you give an example?
*Thread Reply:* And what a fail if we can't even do it properly ourselves 🙄
*Thread Reply:* MUR009.A0201aS0L001R1001.fastq.gz on ENA.. but there is no MUR009.A02..
*Thread Reply:* Yeah stick with what's on ENA
*Thread Reply:* can we have a quick chat.. just to be on the safe side?
So... we are confused. Should we have a single line for each ERR? or one per sample?
What if the library treatment is both udg and no udg. Shall i add both sep by a comma?
As in they mixted the two libraries together in one FASTQ file?
@Megan Michel @ivelsko can either of you hear me?
If there are no library names in either the publication or the ENA table, should I put 'NA' or 'Unknown'?
Is there any library name in the SI or anything?
I have also the same case, where no library name is given for another paper, shall we just use the sample name?
and maybe add numbers if there is multiple libraries per sample?
@aidanva we decided to literally just copy over what is in the ENA table which is unspecified
*Thread Reply:* @James Fellows Yates @aidanva Could I quickly check - if multiple libraries have been used for a single run (ERR entry), and these library names are given in the supplementary information but not in the ENA database, what do we do? Do we keep 'unspecified' as in the ENA database, or do we add the library names separated by ';'?
*Thread Reply:* So all the original libraries have been merged into one FASTQ?
*Thread Reply:* From what I understand, I believe so
*Thread Reply:* Then put unspecieid
*Thread Reply:* Great, thanks!
@James Fellows Yates are you planning to develop a template for uploading data to ENA/SRA that's based on this? So that the information is uploaded in a consistent way for all variables? That would be helpful for both the uploaders and the people putting data here
@channel for the (N/S) americans who are joining, please meet in the big 'classroom' at the top!
@Abby Gancz will you be joining today?
(I think tyou're the last person from the UUS who said they would join)
Can someone check: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/703
Will be back in 20 after :doggo:
Thanks again @channel! We made a lot of progress today, thank you very much for your PRs, reviews and patience while I cleanup a lot of bugs 😬
We plan to run another day in december to try and finish up in the meantime (as most people said they are more likely to dedicate time to it if everyne can sit together in a dedicated event), so I will be sending out a Doodle poll for a second metadatathon in December!
Hopefully we will go even faster next time as everyone is more familiar with the procedure (and maybe we can pick up more people to help out 💪 - so please speak to your colleagues and even PIs 😉 - we can get them all experts of git still).
In the meantime, please keep an eye on review feedback as myself and a few others go through all the ready for review PRs, and if you ever have a spare minute please feel free to do an extra PR here and there!
Note that in the meantime @Maxime Borry refactored the QC check tool a bit to make error reporting easier to parse, so this sort of things will improve the quality of life, so thanks to him!
And please dont forget to list your annoyances found across all the metadata scraping!: https://github.com/SPAAM-community/AncientMetagenomeDir/wiki/Issues-Encountered-when-Metadata-Scraping
AND one more thing (sorry): @channel please label all PRs that are ready, with the 'ready for review' badge. I'm going to look at a couple each day until the next Metadatathon so we can focus on adding the remaining papers rather than reviewing
@channel DOODLE POLL FOR METADATATHON ROUND 2
https://doodle.com/poll/v3aevnu5xi4w3ymf?utm_source=poll&utm_medium=link
💡 PLEASE also share the doodle poll with your collegues/groups, the more the merrier! I will also run the same trianing sessions as necessary (and again across the three time zones!)
@channel last bump for the poll ☝️:skintone2:, please indicate your preferred time ! We have a lot more respondees this time (yay twitter!) so hopefully this is going to be bigger and better :D
I also got through reviews of about half of the PRs, some were good and merged in but some I requested changes so please check your GH noifications!
@channel thanks to everyone who signed up! The AncientMetagenomeDir metadata-thon round 2 will be on Thursday 16th December please put this in your calendar!
I will send more details this week 👍
Details
Thanks for signing up for the second AncientMetagenomeDir metadatathon!
By consensus, this will happening throughout the day on December 16th! Please let me know if you want an invitation to a Google Calednar invite (PM me your google account)
We will follow mostly the same scheme as the first event:
- We will meet on gather.town
- Each timezone will meet and work together, with hand-off events between each timezone
- For each timezone, I will do a introductory presentation introducing both Git(Hub) and AncientMetagenomeDir Please check the detailed information here
The only difference is this time, we will have a team of dedicated reviewers to make the process smoother!
Furthermore, encourage your colleagues to join as well, there is no restriction on the number of participants or level of experience 👍
Please email me or message me on slack with any further questions
P.S. For anyone who tried to deregister from this mailing list, previous emails had a faulty link. Please try again now, and if you continue to receive messages, please email me and I will remove you manually.
@Sreevatshan Srinivasan has joined the channel
So I was right to include the collection year, but be prepared for a lot of 'abuse' in the future 😂
https://www.sciencedirect.com/science/article/pii/S0964830521001724#bib50
🤔 what d ooyu think for #anthropogenic? But I also wonde rif we should drop that table entirely...
@Gabriel Yaxal Ponce Soto has joined the channel
@channel it's me again! You must be so happy to hear that 😉
Just a reminder than we have the Metadatathon round 2 *next week* on Thursday 16th!
Please remind yourself of the schedule and location next week: https://spaam-community.github.io/#/events/metadatathon-dec2021/README. Note the gather.town location has CHANGED.
For those who were in *round 1* please try and check your open PRs and resolve any comments the reviewers have made BEFORE the event so we can focus on new publications!
For those in Asia-Pacific time zones (mostly :flagau: :flagnz: I believe ) please let me know what time you would prefer to start (either the your Wednesday night, or your Thursday morning), as before

Reminder we will be starting TONIGHT for the Pacific/Asia at 21:30 CET
And then TOMORROW at 09:00 for the Europeans!
I will start with a very brief intro for everyone, to also introduce the 'voluntold' reviewing team of @aidanva @ivelsko and @Alex Hübner, and then I will proceed with the the intro to github/ancientmetagenomedir for new comers etc.!
I'm on gather.town for the next 30-45 minutes for :flagnz: :flagau: etc.
@Maxime Borry - @Meriam van Os found the first bug in the new version of 'DirCheck 😆 (or rather, the error has changed)
If a line has extra tabs at the end (basically 'empty' columns) we get a PANDAS error it seems despite the 'check extra missing columns'.
https://github.com/SPAAM-community/AncientMetagenomeDir/runs/4541321193?check_suite_focus=true
Ok reminder to self and everyone else who participated in the previous metadatathon - specifications that have changed:
- Polymerase: is now based on the first indexing PCR/amplification, not adapter fill in etc (that was my confusion about which step would affect damage). It should also be the name from the manufacturer, not wierd abbreviations etc. It is OK use what is reported in the citated paper (e.g. often people will just cite Meyer and Kircher - which is then
Phusion Hot Start High-Fidelity DNA) - Sequencing center: should just be what is reported on the ENA table, we should not fall back to the place where the aDNA lab is (seems to be a lot of confusion here). If the paper and table says two different things, use
Unknown) - readcount: only report numbers in ENA table, not what is reported in the paper (ENA/SRA actually calculates this from the physical file)
- library treatment: Generally assume libraries are none-UDG treated, but if in doubt or multiple libraries merged into one (if they uploaded BAMs), then you can use
Unknown. - Library names: should have spaces replaced with underscores. If there is no library name in the respective column, check also the 'submitted FASTQ' or 'run alias' columns, as you maybe able to view them there.
Reminder:enums must be taken from master not dev!
Otherwise:
Your GOOD MORNING from a frosty Spain! Look forward to seeing European based people in one hour!
*Thread Reply:* Liam is home with a fever today, so my participation will be limited...
*Thread Reply:* Sorry to hear that, hope he feels better soon 🤞
Also: if you've NOT contributed to AncientMetagenomeDir before - please send me your github user ID!
Are any EVA people going to be at the institute at 9? If so I'll go sit in the aquarium/terrarium
@ivelsko @Maxime Borry can either of you hear me? I can't hear you guys?
Maybe you need to go and speak to IT
I will start anyway, for you guys it's mostly new
*Thread Reply:* You're still unmuted while James is talking... 😄
I also can’t hear you nor see anything from your screen
@Arthur Kocher are you in EVA?
@Maxime Borry is going to present the Check tool for any metadathon veterns who skipped the tutorial
(can show later, don't worry 🙂 )
@James Fellows Yates I have another question about the enzyme names. Most end with "DNA" but that's cutting off the name, which is usually XYZ DNA polymerase (DNA polymerase is specified to distinguish it from an RNA polymerase). Should "polymerase" be added back to those names? To me it makes sense to be either the full 'DNA polymerase' or delete the hanging 'DNA'
*Thread Reply:* Hmmm good question.
*Thread Reply:* I think we should fix that after everything is ready
*Thread Reply:* Because we can automate that
Column "strand" does not exist anymore, right? Found it in none of the four TSV headers.
Exactly. So "strand" can be ignored from your mappings table
Yes, I'll delet that
sorry I might have missed it - are people going ahead with adding PRs or are we waiting?
*Thread Reply:* No waiting. Add it and pull it over to the next column in the Project board :)
Sorry for not making that clear 😆
How would you classify a sequencing strategy where they capture human DNA and then shotgun sequence the rest? WGS or...? 🤔
*Thread Reply:* nevermind, I read further and they also do some target capture 🙂
*Thread Reply:* Ok cool!
generally you can just copy over from the ENA for that column
*Thread Reply:* But good you're checking as people don't do that properly sometimes
How specific should sequencing_center be? For example,
Centre for GeoGenetics, University of Copenhagen, Denmark or Centre for GeoGenetics
What does it say on the ENA table?
*Thread Reply:* Centre for GeoGenetics, University of Copenhagen, Denmark
*Thread Reply:* Drop Denmark
*Thread Reply:* but the other centers in the tsv are not as specific, so I wanted to check if I should modify it
*Thread Reply:* Section for Evolutionary Genomics, The GLOBE Institute
*Thread Reply:* Is the other one from the same group... which is probably wrong but too late I guess
*Thread Reply:* Well 'same group' isn't correct because politicis, but you get the idea
*Thread Reply:* https://github.com/SPAAM-community/AncientMetagenomeDir/blob/master/assets/enums/sequencing_center.json
*Thread Reply:* do I add Centre for GeoGenetics, University of Copenhagen to AncientMetagenomeDir/assets/enums/sequencing_center.json ?
*Thread Reply:* Yes, but with a PR made from and requested into **master
*Thread Reply:* Following on from this, should I do the same with 'University of Zurich'? I can't see it in the .json file. So make a PR from the master and requested into the master? Though interestingly, the record on EAN is 'University of Zurich', but the paper specifies 'Functional Genomics Center Zurich' (something to add to the issues encountered file?)
*Thread Reply:* Go with what is on the ENA
*Thread Reply:* Great, I'll make the PR now and go with 'University of Zurich'
*Thread Reply:* Sorry again ^^ I can't commit it to master given it's a protected branch. Would you be able to add "University of Zurich" for me?
*Thread Reply:* Which si your PR?
*Thread Reply:* ahh yes same problem for me ☝️:skintone2:
*Thread Reply:* Speaking of centers, for Ahmed2018 the center name on ENA is Stockholms university (which the paper doesn't state even in the supp files, and the RAW file says NA), json file has SciLife.. What should I choose? 😅
*Thread Reply:* If RAW says NA just put Unknown (or unspecified, or unknown I can't remember... please check the README for what past-James would've said)
*Thread Reply:* Pull Request: Update sequencing_center.json #753
*Thread Reply:* PR: Add Centre for GeoGenetics to seq_center.json #757
Institute of Clinical Molecular Biology is also logged as Institute of Clinical Molecular Biology (IKMB)
*Thread Reply:* Institute of Clinical Molecular Biology (IKMB): KrauseKyora2018, Haller 2021, Susat2020
Yeah, take whatever is on the JSON already (first come serve approach)
*Thread Reply:* No the enum
Martin2013 built libs with two NEB kits: NEBNext DNA Library Preparation for Illumina and NEBNext DNA Library Preparation for Roche/454. I’m not familiar with either, but my Googling suggests the lib prep for Illumina is double stranded and for Roche/454 is single stranded. Does this seem right?
I don't think so... I think it's just slightly different adapters for different machines
That sounds right from their product description
Oh yes you're right I see it now
Yup, I guess!
The pull requests all have changes requested at the moment, so does that mean i can start making new ones while waiting for more to review?
you could go back to ones from round 1 and make the fixes I suggested and double check (not sure if people will come back to them...)
@ivelsko for Brealey you still need to add the qPCR values, correct?
*Thread Reply:* The column name is misleading - concentration is usually a unit per volume, but this appears to want some kind of count
*Thread Reply:* What count? Total reads in library? Number of reads per some volume?
*Thread Reply:* She doesn't have a library concentration, but she has a molecules/volume column
*Thread Reply:* Maybe that's why I'm confused
*Thread Reply:* Let me check
*Thread Reply:* No it is correct I thnk
*Thread Reply:* AL concentration (copies/ µl)
*Thread Reply:* They basically copied what we did for Deep Evo so it sshould be the same
*Thread Reply:* 😬
*Thread Reply:* I made a mistake in the title of the pull request, it's supposed to be 2020 not 2021, so I changed that too
Martin 2013 sequenced all samples on HiSeq2000 and one sample additionally on Genome Analyzer II. I don’t think there is enough info
*Thread Reply:* good question
*Thread Reply:* I would put Unknown
discrepancies between paper and ENA: which takes precidence?
*Thread Reply:* Depends, but normally put as 'unknown' if they are conflicting
e.g. KyoraKrause2018b makes no mention of HiSeq 3000 data, but the ENA tags say otherwise
Do they report a different machine?
*Thread Reply:* Put unknown then
*Thread Reply:* Did yo mean HiSeq 4000 though?
*Thread Reply:* https://www.ebi.ac.uk/ena/browser/view/ERR1883928?show=reads
also think I'm muted on gather.town, tried to speak to the group twice !
I'm being called for lunch - so I will be back in 30 minutes to continue 😄
Feel free to post questions here for me to come back to
Papers which include a mixture of pathogen and host data: how to handle?
*Thread Reply:* Only pathogen
*Thread Reply:* You can just close it, or just edit the current branch to remove the host one. It will update for you - PRs are not static
Reminder: please make sure you open your PRs into dev NOT master, otherwise the aut checks will not execute!
EMEA timezone people - please move into the upper room or the bar (or w/e) before 14:00 CET, otherwise you will get to hear the tutorial stuff again 😄
For people in N. America (i.e. :flagca: :flagus: :flag_mx: etc) we will be starting at 14:00 CET with the intro and tutorial!
@Ophélie Lebrasseur don't forget to go back to your old PRs from the first event: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/677
*Thread Reply:* And also #712
*Thread Reply:* Yes! Thank you for reminding me! I've had to run away for some urgent lab work but should be done and back in 30!
Likewise to @ivelsko for PR #685
@Anan Ibrahim you still have Lammers2021 to correct
*Thread Reply:* Yup I'm joining u guys again later in the day
Slightly unclear why Krause kyora2018b libs PR is failing
*Thread Reply:* and sorry for not pulling my weight more - working off a crap laptop and unsure about PR
Yeah, hold up guys for a second, there was a PR accidently merged in when it wasn't ready
I'm going to fix that now 👍
I've fixed it, I'm going to restart tests now, so please check your PRs in about 2 minutes
Oops, wait one more error sorry!
hey @thedir-metadatathon-reviewers -- the study on parovirus has no actual samples linked on ENA -- but has the algined reads on a github repo... thoughts on how to proceed? ENA: https://www.ebi.ac.uk/ena/browser/view/PRJEB26712 github: https://github.com/acorg/parvo-2018
I assume just sending a link ot a github for a given project isn't what we had in mind.. 😞
*Thread Reply:* Please skip that, I still haven't worked out a way to incorporate this in yet
I think I've fixed the errors now, I'm going back and updating the open PRs
might reuqire some manual work, sorry guys
Ok so: @Kelly Blevins you have your 'own' error now
@Nihan D Dagtas I still have to fix yours
*Thread Reply:* No no, not your problem
*Thread Reply:* An ealrier PR was accidently merged
*Thread Reply:* But you branched while it had gone 'unnoticed'
@Ophélie Lebrasseur I've fixed Ferrari I think
*Thread Reply:* Yes, it's now an error from your data 👍
Ok please proceed - I will kepe monitoring
I think I fixed all the errors now! I'm going to quickly tell Aida how to fix when I do the intro for the Americas
Btw @ivelsko @Nihan D Dagtas @Andrea quagliariello - unless you want to listen again to the presentation I recommend sitting elsewhere 🙂
*Thread Reply:* No problem!
@Betsy Nelson @Christina Warinner @Abby Gancz @Shreya @Sreevatshan Srinivasan (? maybe? don't remember time zone) @Miriam Bravo @Olivia
*Thread Reply:* I have a meeting this morning but I’ll try to come by later!
Woudl be starting about now 🙂
One of the papers used 2 different polymerases (Platinum Pfx and SuperFi). Should I put 'Unknown'?
*Thread Reply:* Which one is used for the initial indexing amplification?
*Thread Reply:* Usually there are two amplification steps during library construction. The first usually uses a non-proofreading enzyme for initial indexing, and the second uses a proofreading enzyme for library completion. You want to record the initial indexing enzyme because that is the one that determines the level of damage in the downstream sequences
*Thread Reply:* They used different enzymes for diff samples it seems. "Platinum Pfx was used indexing PCR for most samples, but since this was discontinued in 2018, Platinum SuperFi was used for some samples..."
*Thread Reply:* Ahh. Do they specify which samples were processed with which? If not, then put Unknown
*Thread Reply:* They unfortunately didn't. Unknown it is.. Thank you!
@James Fellows Yates accidentally committed changes to dev branch rather than my study branch -- what do?
*Thread Reply:* I will also be joining later so maybe I’ll catch you there Miriam:)
*Thread Reply:* 😄 I hope to see you later Che!!
Ok I'm going to fix @Ian Light’s problem now - everyone can go back to the main seminar room and help out the new people 😄
*Thread Reply:* @Ian Light what exacly happened?
*Thread Reply:* i just pushed a commit to dev branch on the .tsv
*Thread Reply:* see https://github.com/SPAAM-community/AncientMetagenomeDir/commits/dev/ancientsingleg[…]ated/libraries/ancientsinglegenome-hostassociated_libraries.tsv
*Thread Reply:* Oh sheeeeet
*Thread Reply:* will revert those two
*Thread Reply:* ok reverted
*Thread Reply:* You'll need to reconstruct it from the old git history
*Thread Reply:* I am getting an error message related to Wagner2014 during the pull request, do I have to do something to change that?
*Thread Reply:* Shit sorry: @Bjorn Bartholdy you'll need to reopen your PR from Rascovan, I made a booboo there trying to fix the Wagner mistake
*Thread Reply:* it also looks like some other committs in the dev branch did the same thing i did..
*Thread Reply:* (look at committs from today)
*Thread Reply:* @James Fellows Yates by reverting the merge commit?
*Thread Reply:* No don't worry: Aida will 'review' directly on dev
@channel I'm trying to fix the Wagner issues, please ignore those errors for the time being
I have to skedaddle. I will contribute off and on when I can, now that I know how! Thanks for the great overview of github and the ancientmetagenomdir, @James Fellows Yates(both times lol)!
*Thread Reply:* Thanks very much!
*Thread Reply:* I second Kelly. Thanks a lot x2! @James Fellows Yates
ok I THINK all the Wagner errors have been fixed now, please ping me here if you get errorson it
*Thread Reply:* just opened a PR for the wagner data -- not sure if the automated checks ran. i will check on this later tonight or tomorrow!
Thanks for all your help and organizing this James!
@Maria Lopopolo @Gabriel Yaxal Ponce Soto please remember to open your PR into dev not `master!
@Ian Light your secondarysampleaccession is incorrect, you should have a single SRS per run in this case
@ivelsko I think you put this in the wrong place: should've been master
Ah you also did it on master, nevermind!
So question, for Dux2020. They do an a "Inverse rRNA selection", so @Iseult set this as "RNA-Seq" as library_strategy. What shall I add in the enums?
Good question - what does the ENA say?
Ok let me check the ENA API
These are our options: https://www.ebi.ac.uk/ena/portal/api/controlledVocab?field=library_strategy
I will then go for RNA-Seq, what do you all think?
Context: " To retrieve MeV genetic material from this specimen, we first heat-treated 200mg of the formalin-fixed lung tissue to reverse macromolecule cross-links induced by formalin and subsequently performed nucleic acid extraction (17). Following DNase treatment and ribosomal RNA depletion, we built high-throughput sequencing libraries and shotgun sequenced them on Illumina® platforms."
none of the other RNA-Seq does really apply....
@channel remember that for the sample_acession column, you should only put a single SRS
Not the long list of comma separated ones, when people did that wrong
(there are two columns in the RAW table)
(one from ENA one from the 'Dir sample metadata table)
Errors with ancientMetagenomeDirCheck like the one below - how do you dig into underlying issue?
Always check the the comment from github-actions-bot on the main conversatoin tab
I shouldn't have shown everyone the checks tab, sorry 😅 that is just for nerds 😬
had a lot of fun but unfortunately I have to go! happy to help out in the next one or if this is rolls on past today
Ok @channel! I need to leave now for a while.
*Thank you* to everyone who has contributed to day, we made really good progress, and are now over half way, there is only about a third of the publications that have not yet been started! All your hard work (and patience with the bugs :D) is much appreciated!
@Maxime Borry has reported he *almost has an finished alpha version of the tool*, so we will work out how to introduce that to everyone later.
*I will come back* to check in on people from the Americas at about 21:00 CET (but feel free to message me on slack if you want with Qs).
We will need to do *one more event* early next year I think, so if you're interested in joining for one(?) more to try and power through the last set of publications - please leave the emoji 🐒 on this comment so I can guage interest
and of course feel free to keep working on these outside of the events (at least making your corrections after review would be really helpful)!!!
Thank you so much @James Fellows Yates for your patience 😇
You're welcome 😄
also next time I'll be organised and do a proper 'checkout' session... I keep forgetting to do that. Sorry!
You also get the award of the quickest running avatar!🏃♂️
I'll be back in about 1h @I-Ting Huang, @Olivia, @Christina Warinner, if you want to talk about or go through anything
lol closed the tab accidently, one moment
@channel Once again thank you much for all your efforts yesterday! I would like to give a few stats, and also ask for some feedback to make next event(s) better/work smoother!
To give you some insights of the scale we are working, we added since the previous event:
• Single genome host associated (pathogens): 239 -> 652 (+413) new libraries • Metagenome environmetnal (sedaDNA): 47 -> 69 (+22) new libraries • Metagenome host-associated (microbiome): 484 -> 545 (+61) new libraries For a grand total of 496 new libraries (note this is slightly biased by the order of the issues, with single genomes being top of the project board)
We also have started work on almost 2/3rds of the publications, which is great progress!
So 👏 🙇 to the 27 contributors who joined this time around!!
I would be very grateful if you have a spare 2 minutes (literally) to give feedback here: https://forms.gle/2GWFJax25yWm8ygo8 I will make sure to make any future events run more smoothly!
I will spend the rest of the day doing reviews of the open PRs to try and close some more. Please continue working in your spare time, and base d on the feedback form, I will start arranging a third event if we feel it necessary!
Thanks @Alex Hübner for reviewing Warinner/Fellows Yates!
My PR #800 got an error (This branch has conflicts that must be resolved) and doesn't proceed to the validation step. What should I do?
*Thread Reply:* Ah, basically dev was updated since you made your branch, and you have made changes in the same place as the update. Git doesn't know which changes to pick, which is why it says that.
It's normal :)
I'll resolve it for you in 20m or so!
@Nihan D Dagtas conflict resolved and tests running 🙂
Btw I've now found the settings to restrict merging to only once a reviewer approves the PR, and restricts merging also to the reviewing team to make sure we done have the same accidents like last time! So people can feel a bit safer that mistakes won't occur in the future 🙂
@James Fellows Yates I checked the previous threads about including negative control samples but wasnt sure. Shall we add them as samples in the libraries dir?
No we do not include negative controls.
(couldn't find a nice way of including them at the time)
@channel ☝️ opinions plz! @Maxime Borry’s filtering and downloading tool is almost ready, but we need a new name for it (as it's not just checking stuff now, and AncientMetagenomeDirCheck is long and clunky).
So please vote, and add your own suggestions!
So I'm just getting back to reviews after a few days, and whooooooa! Thanks to @Raphael Eisenhofer @Nihan D Dagtas @Anan Ibrahim, and in particular THE BEAST that is @Kevin Daly
I promise to get to them ASAP, I will at least get throug hthe maintianence PRs for sequencing centers etc today
I really appreciate the continued work on this! I think at this rate we will definitely able to complete everything in January/Feb next year 💪
Also sneak-peek on the awesome work @Maxime Borry has down for the download tool 😎 👇
As a reminder: if you're updating enums, please do this on PRs forked off master NOT dev (awkward, but it's the way thedirCheck has to check against web versions of the enums)
Prepare your inboxes 😬, I've got an afternoon just for reviewing!
@Shreya https://github.com/SPAAM-community/AncientMetagenomeDir/pull/724 I've already reviewed this one (still unresolrved) - we did we talk previously about this paper? Is there a reason why there is only one library, but 4 strains were reported in the paper?
*Thread Reply:* we did talk about it! long enough ago that I don’t remember what we discussed… will take a look later today
So, lots of distractions today, but:
@Shreya @Adrian Forsythe @Iseult @Ophélie Lebrasseur
I requested some changes (minor in most cases)
But i merged in a couple from @Andrea quagliariello @Jasmin Frangenberg!
@Kevin Daly I closed the Illinois PR because there is a discrepency between the paper and the ENA/SRA metadata, so I suggest updating Witt to just 'Unknown' for sequencing center
*Thread Reply:* sorry for missing that - but i suppose that's what the second pairs of eyes are for - when I checked the witt2021-libs branch, "Unknown" was already used as the sequencing centre - does PR #805 just need to be reviewed one final time and/or accepted?
*Thread Reply:* I updated it for you. I still need to review it properly, so you can leave it for now (I'm going from oldest to newest), but wanted to let you know why I closed the Illinois PR and why your PR would've looked different
OK! 72 issues still open (only 35 not started) and 87 DONE! We are over halfway 😎
I'm going to keep picking at away at the reviews over the next weeks, sorry this is taking such a long itme
Is there anyone on the reviewing team who could quickly approve this one?
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/825
@Kelly Blevins I've also left some questions for your PR for Maritn2013
*Thread Reply:* Mostly minor but there were so many rows I may have not been in the mood to update it myself after going through so many other PRs... sorry - nothing personal 😅
*Thread Reply:* wasn’t grimacing at you. I was grimacing at myself! Should be fixed now (after spending an hour re-learning everything I learned in December and then promptly forgot).
*Thread Reply:* No worries - was just giving clarification that it wasn't a major thing 😅
But I've closed more from @Andrea quagliariello and @Iseult - thakns very much!
38 people 😱 Shout if you don't like your photo - I stole it either from Github/Tiwtter or a Google Imsages search
*Thread Reply:* Not a photo issue, but it looks like Raphael's name is split on 2 lines - the 'l' is on a 2nd line
*Thread Reply:* No problem with the photos, but the Irish flag is green white orange rather than the other way around 😋
*Thread Reply:* Uhh oh! That's quite a big bug in the Google emojis then 😱
*Thread Reply:* I thought it looked a bit odd but thought I was being silly
*Thread Reply:* Ooooor i accidently selected cote de ivorie 🤦♀️:skintone3:. I will fix tomorrow 😅
*Thread Reply:* I think I must have searched with i and was to fast and selected the first hit
*Thread Reply:* Sorry 🤦♀️:skintone3:🤦♀️:skintone3:🤦♀️:skintone3:
*Thread Reply:* My Photo looks a little wider than normal... Not sure. If you didn't play with the aspect ratio at all, nevermind. Hmm some others also.
*Thread Reply:* I think it's a artefact of the cropping unfortunately...
But I don't think people would study it so closely for it to be an issue. But if you don't like it let me know I'm happy to try another trick
*Thread Reply:* Hello ^^ Thank you for checking @James Fellows Yates! I'll send you more of a portrait than the one I currently have. Also, would you be able to add the Argentinian flag? You wouldn't have known, but I'm half based in France, half based in Argentina 🙂 Ta!
*Thread Reply:* Tha'ts awesome! absolutely 👍
*Thread Reply:* Wombat 🤣
We will need ot do a cleanup at some point, I'm confused as we seem to have an extra 10 library issues still open, that aren't listed on the project board
But we are down to 16 open pull requests! And work has started on all but 33 publications!
WE ARE GETTING CLOSE PEOPLE
@Bjorn Bartholdy I changed my mind about waiting to use your PR as a guinea pig, lets get it in now 👍 did the review for you: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/634#pullrequestreview-882565535 And I'm already talking separately to @Arthur Kocher about his PR for Kocher 2021
Once they are merged in we must remember to make issues to add the libraries for those two publications too
*Thread Reply:* When will be the next release?
*Thread Reply:* Even we get the libs done, or do you want it earlier?
*Thread Reply:* I'm in no rush, not sure when I can get to it, but I also don't want to hold up the next release
*Thread Reply:* Ok, if you don't have time let me know and I can correct for you
*Thread Reply:* @James Fellows Yates I'm hoping to get to it by the end of this week, but feel free to make the changes if you also find some time!
*Thread Reply:* This week is fine
Ok, so we are down to ONE non-problematic still-open PR! Thanks very much to @Alex Hübner for his help powering through all the reviews.
Once I've resolved the remaining problematic ones I will do a call for (hopefully) ONE FINAL PUSH to get the remainder of the publications in (hint: most are single-genome publications which normally equate to a single sample, so should be fast ;)). So please have your calendars ready for one final metadatathon! Thanks again everyone!
*Thread Reply:* Thanks to you @James Fellows Yates for all the fantastic and hard work!
@channel I need some input/advice/suggestions on how to deal with an awkward publication for the pathogen table.
Lugli et al. 2017 reused data from Maixner et al (2016, Ötzi's H. pylori publication) to recover additional genomes of other microbes. So we list it in the single-genome samples table, as it represents distinct genomes.
HOWEVER, my question is how do we include this in the library/run table?
Should we keep these entries, but results in mostly 'duplicated' library/runs as what we already have with Maixner (making it more difficult to do stats, e.g. no. of libraries), and risk of people not citing the original publication. OR Do we discard these and rely on people to know to search for ERS codes in the library table, once they've selected the samples they want from the sample table?
Based on how AMDirT works at the moment, I think the former would be preferential? But equally when downloading this could result in redundent libraries...
Feel free to give your input as to how you were search for data (AMDirT implenentaiton is a specific question to @Maxime Borry)
isn't in our checklist that the data should be newly sequenced metagenomes?
Yeah, but the single-genome one is always special 😉
Because you can have multiple generated genomes
(metagenomes you can't really create new sequences)
that's it true... but then I will say we keep it and just make people aware that duplicates can exist
and when doing starts people should be removing duplicates
it may also be interesting to keep in, since people may be interested on how many papers have been produced from a single dataset
I agree, I would keep the entries in the library/run table also for clarity purposes. I tend to feel confused/uncertain when trying to identify such information but the information isn't provided and I have to do some cross referencing. I rather have the data duplicated but clear.
In addition to the above: I would like some second eyes on this one - https://github.com/SPAAM-community/AncientMetagenomeDir/pull/806
I've updated it how I tihnk it should work but I had to jump across many different tables and tabs so I might have made copy paste errors (@Alex Hübner, @aidanva or @Åshild (Ash))maybe?
@channel IT'S ME AGAIN! WITH ANOTHER DOODLE!
For the LAST AncientMetagenomeDir library metadatathon! This time for either the second half of March and April.
Please fill in here with your *full name*
https://doodle.com/meeting/participate/id/lejJpRWe
Once we have finished this, we should have all the PRs in, and we can finish @Maxime Borry’s downloading tool and we can start preparing a publication!
*Thread Reply:* Hello @James Fellows Yates - Just filled in the doodle - all the dates in April for me are provisional as, as you know, I don't quite know where I'll be then, but I'll do my best to make it 🙂
*Thread Reply:* Also - argh I can't go back to editing my name so it's my full name 😕
*Thread Reply:* Don't worry, you i can recognise easily 😊
*Thread Reply:* I will join this time if the date that is decided on works with my schedule :)
*Thread Reply:* that would be really helpful 😄
@channel last call ☝️ poll will shut tomorrow
@channel (sorry for the millions of @channel)
Date is set: April 6th 09:00_16:00 CET (and early start/late finish for people in Pacific-Asia/Americas timezone!)
Please add it to your calendars!
@channel This is the week! Lets finally wrap up the 'big' library metadata conversion of all existing entries on Wednesday 💪
For those in *pacific-related timezones* (basically anyone getting up earlier than CEST) (or people who are really enthusiastic 😉 and want an extra day to work on tihs) - we will be meeting on your afternoon of April 5th to get you going, and allowing you to work over the European night.
Therefore for the *pacific timezones*I will be on the SPAAM gather.town on April 5th, 08:30 CEST or (So ~18:30 NZL, ~16:30 AUS, ~15:30, KOR) to do the introduction and then I will hang around to help out for a bit.
Then at same time April 6th I will join again to catch up with you guys, and you can also join to meet the peepz in EMEA timezones 👍
Then for *EMEA and Americas* we start on April 6th
EVA people: we will meet in ABI Seminar room!
I'm ready for the Pacific TZ start!
https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
@valentinav I will do a quick review now for you!
@Alex Hübner I removed the sample-level issues from the project board, this was confusing for first timers to the 'Dir. we can add them again if we get through all the the PRs tomorrow - but will make sure to make it clear it's a different type of PR
*Thread Reply:* Sounds good, I agree.
Ok @James Fellows Yates , will be there when the next meeting starts tomorrow. Cya
@channel starting in 5 minutes: https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
Slides: https://hackmd.io/@jfy133/HyKgEJVLY#
Hi, I've a quick question- if the publication only mentions paired end sequencing but the ENA tsv has both paired and single in the library layout column, should we go for what the ENA table says?
*Thread Reply:* Scusi!?!
*Thread Reply:* Can oyu send a screenshot?
*Thread Reply:* Ah ok, yeah in this case trust the ENA table
*Thread Reply:* "Amplified libraries were pooled with other indexed libraries and sequenced on Illumina platforms using the Paired-End mode. Screening and library validation were carried out on a MiniSeq instrument (2x80 bp reads) at the CAGT laboratory. Libraries showing endogenous human DNA content higher than 10% were selected for deep-sequencing and sequenced on a HiSeqX (2x150 bp reads) at the CEA/CNGM (Evry, France) or on a NovaSeq S4 (2x150 bp reads) at SciLifeLab (Stockholm, Sweden). These represented 5/12 and 7/11 remains for Mont-Aimé hypogea I and II respectively, 1/6 samples for Mas Rouge, 4/7 samples for Grotte des Tortues, 3/7 samples for Grotte Basse de la Vigne Perdue and 6/10 samples for Grotte du Rouquet."
*Thread Reply:* omg that's rzy 😆
*Thread Reply:* Can you send me the PRJ code?
*Thread Reply:* I want to double check something
*Thread Reply:* Ok give me 2 mmutues
*Thread Reply:* Checking now
*Thread Reply:* Go with what's on the ENA
*Thread Reply:* it corresponds to what files are available
*Thread Reply:* Unless they have uploaded both raw AND merged reads 🤔
@valentinav if it's just the calculus sample: https://sid.erda.dk/wsgi-bin/ls.py?share_id=E56xgi8CEl;current_dir=.;flags=fal
@Raphael Eisenhofer I'm going to have review the santiago rodriguez PRs when I have a bit more time, I need to find out what each MG-Rast accession code refers to and I can't easily find that in the docs
@channel I'm going for lunch now, if you want to see a demo of the tool please reconvene by around 13:00 CEST and @Maxime Borry and @Nikolay Oskolkov will show this maybe around 13:00_13:00 ish
@Maxime Borry @Nikolay Oskolkov demo at 13:30 ok?
@Raphael Eisenhofer we will have to remove the entire Santiago Rodriguez (at least) 2017 dataset
both from library and sample
The raw reads aren't available
https://www.mg-rast.org/mgmain.html?mgpage=download&metagenome=mgm4629032.3
*Thread Reply:* The first set of data we can get to is step 5... the output from bowtie2
You can only get host-removed reads, but in FASTA format (i.e. all quality scores removed)
@Alex Hübner what do you think about this? Is it fair to remove ? Or should we keep?
In the pathogen table we allow more variation, because people upload ocnsensus sequences
but in the 'microbiome' table we've been much more strict
Would this also apply to 2016?
I think we could keep them still. There might be something interesting in them and it would be up to the individual scientist to deal with the missing base qualities.
But yes, these data are only FastA files, too.
Mmmm ok will need to think how to flag that... I guess we can add a file_format column or something I gues...
@channel Maxime is going to demo the download tool now
So you have 2 minutes to join if you wanna see 😄
@Alex Hübner @aidanva @Åshild (Ash) thoughts?
*Thread Reply:* Did Mühlemann et al. upload to GenBank or to ENA?
*Thread Reply:* but they still generated the sample upload sheet 🤷
*Thread Reply:* So they have ERS codes, and these ARE linked to the genbank thing
*Thread Reply:* What is the question here? whether to add ‘Unknown’ in cases of lack of Project Accession?
*Thread Reply:* Yes, and whether it's ok to use the Genbank ID for both sample and run accession
*Thread Reply:* dad joke
Hi @aidanva and @Åshild (Ash). I have many questions about how to handle Worobey2016. The samples were not generated using NGS; amplicons were capillary array sequenced. The available data are consensus seqs in one NEXUS format. I am thinking that all columns should be NA except projectname publicationyear publicationdoi samplename archive archiveproject sequencingcenter filetype downloadlinks. I am not sure about library_name, because the seq data have names that are different from the sample names, but they are not libraries.
*Thread Reply:* Hi @Kelly Blevins i had similar issues with BarGal 2012.. maybe you want to have a look how i managed it there.. (with lots of help from James)
*Thread Reply:* not exactly the same, but some columns could be similar.
you are right, if you don't have any of the information, just put NA/Unknown depending on the column
@Arthur Kocher was having a similar problem where he only has consensus. If you can find a download link then include it, if not put as NA. When making the PR, mark it as problematic
should I include the sequence names under library_names? or just leave them out entirely?
do you know which correspond to which sample? is it easy to infer from the paper?
yeah. the sequence names contain more metadata than the sample names. For example, consensus seq 1979|B|US|1979|1NY79 came from sample NYC1
Hmmm, I see your point. Either you leave it out, as it is strictly not a library name, or we add some extra info to the library_name column that specifies that this can also be sequence name in the case of consensus sequences. @aidanva what do you think?
I would leave it our
One more question. Most of the enums only have “Unknown” as an option. But “NA” or “Not applicable” is more appropriate for Worobey2016 for all of the NGS-centric columns. Should I add an NA option to the enums?
*Thread Reply:* Follow README as much as possible, NA is a special thing in JSON so we can only use it with Numeric columns
BarGal might be a good example like Gunnar said. Try and do as much as you can, and then label it as problematix and I'll work out what to do.
I have got an error during my checks which I don't quite understand:
I'm not quite sure where that matching is coming from?
Remove the doi.org/ at the beginning
Btw @channel for anyone in the Americas still going, I'll be available in about 30 minutes if there are any outstanding questions or problems
*Thread Reply:* All good on my end, thank you 😄
*Thread Reply:* This American will probably show up long after all of you are asleep
*Thread Reply:* @Shreya if you could check if you have any remaining from previous ahckathons not finished that would be great ❤️
Ok 30 More, need to wait for maia to fall asleep 😬
*Thread Reply:* Just a note - I'm the only one in the space so I wouldn't stress too much ^^
*Thread Reply:* Is there anything else I can do if all the 'To Do' have been done?
*Thread Reply:* Nope! Well yes (we have some new sample level metadata to add), but I consider the main objective completed which is fantastic!
I would say take the extra free time for your own stuff instead
Ok, if no one is around then we can just wrap up here
@channel thank you VERY VERY MUCH for everyone who has contributed today, and the previous two hackathons
128 publications COMPLETED! Which is FANTASTIC! And just a few stragglers left 😄
Myself and a few 'hardcore' people will wrap up the remaining ones as these are a bit more tricky, but thank you thank again for all your hard work. Like I keep saying, this sort of project only works when you have a critical mass of volunteers and you've made this a success!
Once we've finished off the last few, we will focus a bit more on getting the release ready and the last bit of polish to the tool from @Maxime Borry.
at this point I will be in contact with everyone who has made a PR to start planning a publication
So keep your eyes peeled on this channe land also via email.
And also remeber: please don't stop here, keep monitoring the repository and add ppublications when you see them coming out to hte issues, and making PRs - this sort of thing can maybe take 30 minutes a month for a group of 10-20 people and still having it ticking along nicely and act as a really important and useful resource for the community
Thank you again ❤️ ❤️
OK @channel before you take a complete break - for everyone who was there yday, please go back and check all your open PRs and whether the auto-checks have passed!
there are quite a few that still have errors. Always double check the README in the library directory if you're unsure and/or don't undersand the error the bot is reporting
To help the reviewing team I'm going update labels so only those with passing checks are 'ready for review'! So please update that accordingly
I will be going through all the remaining/problematic reviews next week!
Ok... so Santiago Rodriguez, MG-Rast doesnt even have the host removed reads, no md5sum (compared to previous), and odwnload sits there fore ever.... @Alex Hübner Drop?
Yes, let’s maybe just drop it then.
OK! Thanks. Sorry @Raphael Eisenhofer - I'll remove everything
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/879
Should we rename _libraries to _data level metadata everywhere... was just thinking particularly for the single genomes many are cosensus sequences which aren't really libs anymore...
Respond 1️⃣ for agree and 2️⃣ for disagree
Comments can go in a thread 🙂
@irinavelsko @Alex Hübner @aidanva and particularly @Åshild (Ash) it would be nice to get some feedback on this: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/668/
It's PCR data of genes of Flu Virus. I've added how I think it should be added, so it would be good to get a review for this other than myself
(only 7 entries, one sample)
And Ash (who I just saw is on holiday), for Mendum, I've tweeted TreeBASE asking why the website is locked... https://twitter.com/jfy133/status/1513836303946821632
gonna ask there. But their github has not been updated since 2018...
I think it might just be worth removing that sample
Let me know if you don’t get a response to your email, and I can email the authors to ask them to release the raw data
We should probably ask that already tbh
The TreeBASE files arne't that useful anyway IIRC
But I will wait to email after the easter holidays
@Åshild (Ash) @s.wasef @Nihan D Dagtas @Bjorn Bartholdy @Olivia @Ian Light @Jasmin Frangenberg a little nudge that you still have incompleted PRs to add! I'm getting close to running out of problematic PRs to review, so new ones would be welcome
Thanks @Nihan D Dagtas and @Ian Light!
I'm now down to the Bieker stuff and that will be all open PRs, so I will be available for somemore reviews!
note I have two house kepeing PRs for the reviewing teams (@aidanva@Alex Hübner@irinavelsko)!
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/879 https://github.com/SPAAM-community/AncientMetagenomeDir/pull/880
Two from me I can't review myself 😬
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/892
We are getting really close to a release now I think!
Then @Maxime Borry can finalise AMDirT and we can prepare a manuscript!
For ‘sequencing_center’ in the library dir, is this the same as ‘center name’ on ENA or ‘Submitted by’ on SRA?
*Thread Reply:* If it's not clear just put Unknown
*Thread Reply:* I might drop that column as it's often a need
*Thread Reply:* Also, sometimes people send samples for commercial sequencing, which is what we do for everything
*Thread Reply:* There is no ‘sequencing_center’ choice on ENA, just wondered if this the equivalent to ‘center name’
*Thread Reply:* > Also, sometimes people send samples for commercial sequencing, which is what we do for everything > This is also why I'm thinkibg of dropping
*Thread Reply:* Maybe replacing with codes from ancient-metagenimic-labs
*Thread Reply:* As it's the extraction/library prep which has a stronger bias than the sequencing in most cases
*Thread Reply:* I think that is a much better idea, although can get complicated when samples are generated in multiple labs…but useful to know
*Thread Reply:* I might be getting a Hiwi that can help with such things 😏
Would anyone be willing to review this and see if we are missing anything: https://www.mdpi.com/1999-4915/14/6/1336/htm
Table 1

Last environmental library PR !
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/899
@channel we are REALLY close now! https://github.com/SPAAM-community/AncientMetagenomeDir/pull/857
Just need reviews!
Then that's all the library metadata for all rpeviously published libraries 😄
Which is more than FOUR THOUSAND libraries so fantastic work everyoen!!!
Thanks @Alex Hübner! We are down to 3 PRs!
TWO MORE TO GO!
last one
DONE
*Thread Reply:* 🤩 You've earned such a well deserved rest!
I will do some last checks and cleanups, then we can finalise AMDirT with @Maxime Borry and we can start preparing a manuscript!
OK clean up PRs incoming:
https://github.com/SPAAM-community/AncientMetagenomeDir/issues/732
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/903
(these are really simple! Anyone can review 🙂 )
*Thread Reply:* Took care of the first one 🙂
*Thread Reply:* Friday afternoon a good time for review requests I see 😆
*Thread Reply:* Extra points for doing it during USA waking hours 😄
*Thread Reply:* @James Fellows Yates I’ve approved it, do I go ahead and merge or do you?
*Thread Reply:* You can mege
*Thread Reply:* Satisfying button 😎
*Thread Reply:* same for you @Bjorn Bartholdy
*Thread Reply:* (working out the third fix)
OK, I will continue preparing everything for a release next week, but need input first:
- Should we drop anthropoligical metagenomes as there has been 0 interest? Can add back again later in the future. Or better to just leave it and wait for someone to pick it up.
- Library metadata changes:
◦ Rename
archive_run_accession>archive_data_accession(as we have consensus sequences which aren't runs) ◦ Renamelibrary_publication_doitodata_publication_doi(to account for when genome produced from public data, and original paper should be also cited) ◦ Remove sampling date (sample list) as this is difficult to define and rarely reported ◦ Remove nominal length as variable as this has been ridculously inconsistent and mostly NA. I don't think this will influence downstream analysis ◦ Remove sequencening center as also ridiculously inconsistent. We could in the future re-add this column but using codes from AncientMEtagenomeLabs as I think the aDNA lab woudl be a more important confounding factor in analysis
Please ✅ if you agree with all, and if any queries/isssues/questions please add in a thread below the list of suggestions
@channel ☝️ the more feedback the better!
*Thread Reply:* I'm particualrly looking at you @Alex Hübner @irinavelsko @Åshild (Ash) @aidanva as often the most opinioanted ;)
*Thread Reply:* I'll make a PR into dev with the proposal in a bit if it helps
Furthermore please also put forward any other suggestions you may have 🙂
Here are my two cents:
- Yes, there are no samples.
- Regarding the renaming, I am fine with it because we would be more direct about it. For removing the nominal length and sequencing center, I agree with you, too. It’s inconsistent and doesn’t really help. Adding a more specific column like the AncientMetagenomicLabs might be more helpful in the future to discover batch effects.
Regarding the sampling data, I have no strong opinion.
Ok, then I've made the following PRs for review:
Remove anthropogenic table: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/904
Remove unnecessary /rename columns:: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/907 (note this looks worse than it is, my auto-formatter for YML/markdown files had a lot of fun apparenrtly)
Once they are approved and in I'll do the last validity check we are not missing anything, and generate some new stats tables and I think we are ready for a release 🙂
I agree with all the above suggested changes. Can you remind me which column “nominal length” is a variable for?
Sorry, that was for sequencing_cycles
(I forgot I renamed in for the Dir)
I also agree with all the states changes
Any takers on teh removal of anthropological meagenome: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/904/files?
*Thread Reply:* me, me!
*Thread Reply:* Basically check any reference to anthropological stuff is gone other than the note in the README 👀
Another tidyup PR!
I basically simplified the assests directory (where we place all the images/scripts etc)
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/908
Ok now a tiny correction to Fotakis 2020: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/909/files
*Thread Reply:* Serves me right for manual editing!
*Thread Reply:* 2 minutes...
*Thread Reply:* But the info should now be the same as here basicaly: https://github.com/SPAAM-community/AncientMetagenomeDir/blob/7e09f7d5b69cc71afcff4649d4d0ebdbd20c1a07/ancientsinglegenome-hostassociated/libraries/ancientsinglegenome-hostassociated_libraries.tsv#L1089
*Thread Reply:* Ok try again 🙂
*Thread Reply:* Gotta love those rogue spaces at the end of a line 🤣
*Thread Reply:* Thanks very much @Bjorn Bartholdy!
*Thread Reply:* Back to making silly R plots
OK me again @channel (yes, I have a mostly meeting free week this week 🎉 sorry for the :spam_dance: )
I need a review for: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/908
This adds new summary R scripts and figures based on the library metadata. I'll only diplsay the culmulative reads and libraries on the main README, but out of curiosity I added the same library plot but grouped with different other categories.
Please let me know of any general feedback, or any further plotting requests!
OK one sec... slack won't display SVGs anymmore
*Thread Reply:* Is the third one (libraries per year grouped by layout) showing what it's supposed to be showing? I don't see the groups.
*Thread Reply:* No it's not, good catch 🤔
*Thread Reply:* Meant to be showing paired or single
*Thread Reply:* There also seems to be a mismatch between the standard cumulative plots and the grouped cumulative plots. They should contain the same number of publications, but the grouped plots seem to be missing some. The grouped plots also don't seem to be displaying the cumulative sum (right?)
*Thread Reply:* As for general feedback, it's definitely a nice way to see various trends over time!
*Thread Reply:* I just realised this morning that the group by summary may not include sets if something is NA... I'll go back and check
*Thread Reply:* Ah no it's the joining operation not making (obvs) categories with 0 when that particular year doesn't have any data
*Thread Reply:* 🤦♀️:skintone3:
Pre-release checks summary: only missing 2 publications, and 7 with issues (but mostly because they were difficult because teh data was uploaded wierdly), so congrats everyone!
@channel OK, we now have 14 'clean up' PRs to clean up minor mistakes (typos, accidently cleanup libraries).
I would be very grateful for reviews on these - they shoudl take you 5-10 minutes absolute tops in most cases as I've described the issue/solution in most of them and normally consist of only fixing 2-3 lines 🙂
I'm happy to assign people directly if you prefer that method 😉
Once all in I will do my table discrepancy check one more time, and then basically we are ready for release!
*Thread Reply:* please just assign me to some, i’ll have time later today!
*Thread Reply:* Assigned you 3 pathogen ones!
*Thread Reply:* Assigned! More of a mix this time but should be pretty simple
*Thread Reply:* Now you have 12, @James Fellows Yates. ;)
@channel reminder that if we don't get reviews, that means no release, which means to no paper 😉 the more people who help out the less work everyone has to do 🙂
https://github.com/SPAAM-community/AncientMetagenomeDir/pulls
*Thread Reply:* I can do some later today - what needs to be done exactly? if you just assign me a couple I'll get around to it in a few hours 🙂
*Thread Reply:* Just check that my changes don't have typos/make sense :)
*Thread Reply:* Here you go:
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/915 (I think you were involved with this set of metadata at some point, basically there were a lot of missing libraries, so it adds the missing ones) https://github.com/SPAAM-community/AncientMetagenomeDir/pull/918 (ez-peasy typo fix) https://github.com/SPAAM-community/AncientMetagenomeDir/pull/930 (typo fixes)
*Thread Reply:* Thanks very miuch @Åshild (Ash) and @Iseult! 6 more lean ups to go 🙂
The only one I would like someone more 'senior' in the project to review is #917, because that's a bit more complicated one
*Thread Reply:* Thank you @Åshild (Ash)!
Thank you very much to @Iseult and @Åshild (Ash) for the flurry of reviews! Just missing two library PRs, and there is the images update PR (see my message above) and one correction PR :)
We are very very very close now
Does PR #908 need to be tested locally or anything before merging?
Not yet, need opinions on the figures here: https://spaam-community.slack.com/archives/C0183TC8B0R/p1657114641355499
If they look ok, if any are not useful, if there are others that should be made instead
Thanks to @Åshild (Ash) @Shreya @Bjorn Bartholdy I've fixed some errors in the figures. Please look again and give feedback! Anything you don't like? Anything you think is missing/would be useful?
*Thread Reply:* I think the plot about what type of sequencer was used would make more sense as a line plot of the usage per year rather than a cumulative sum plot. There are too many categories in the stacked bars to get something meaningful out of it other than the major trends. It is particularly hard to infer anything from the environmental samples because you prefer to use the same y-axis scale and there are the fewest libraries. So I cannot really take much from this plot. The other ones look good.
*Thread Reply:* @Alex Hübner does this look better
@channel it's me again (sorry)
I'm doing a call for people who are comfortable with R and python!
I'm looking to do a little 2 day hackathon to complete the next Dir release (now slated for September - version 22.09).
1️⃣ would also like a few people with R experience to help do a big validation of all the library metadata (making sure we aren't missing anything) [basically - loading tables, doing diffs/filtering/maybe more ggplots]
2️⃣ people familiar with Python to help @Maxime Borry finish the outstanding functionality/testing we need for AMDirT.
Are there any takers? You don't have to be 'experts' - just people who can start exploring right away (i.e. no training for this event)
(@Nikolay Oskolkov @Jasmin Frangenberg for example, as you already helped a little with AMDirT)
*Thread Reply:* (I clicked both, prefer 2️⃣)
*Thread Reply:* I'm in, but not for September, as I'll be in conference, and holidays
*Thread Reply:* Eek. End of august?
*Thread Reply:* I'm only free again from the beginning of October
*Thread Reply:* I am in guys, just let me know when. I can help with both R and Python depending on where needed
*Thread Reply:* I'll send when2meets on monday!
what would be the tentative dates for the hackathon?
Would do a when2meet to see when most people can do it 😉
I would like to help with R but will confirm once I know the schedule better because I am moving to another country in one month :)
*Thread Reply:* 😱 good luck with the move! Could you indicate with the emojis too?
*Thread Reply:* Indicate what? Lol sorry for not getting it 😅
*Thread Reply:* that you would help wiht R
Ok slight correction:
R people in September! Python in October (due to @Maxime Borry being away in Sept)
*Thread Reply:* Cool, I'm down for both then!
OK: so we now have a 'deadline' - @Christina Warinner would like to have a release sooner rather than later for use of the stats for a paper.
Therefore R people who are in this @channel who are interested in sitting together for a couple of hours to check everything looks good for the release (no mistakes, missing anything, better plots etc.): lets see if we can find a date in August so we aim for a release deadline of *1st September*
Please fill in the when2meet - all times CEST/Berlin time: https://www.when2meet.com/?16395836-kvFY6
I'll shut the poll today or tomorrow depending on the number of respones
For context - it's because I'm going to submit a paper soon that includes analysis of the entries in ancientmetagenomedir and I want to properly cite and credit all your hard work!
@channel it looks like the best time for the R validation session is Friday August 26th 09:00_13:00 CEST!
Please mark that in your diaries/calendars - if you PM me your email I will add you to a google calednar invite (if you prefer)
*Thread Reply:* @James Fellows Yates sorry, I missed a conflict in my calendar... 😬 I can still join after 13.00 if necessary!
*Thread Reply:* No worries!
*Thread Reply:* @Olivia will likely be joining around the nanyway
@Olivia I know this will be a bit early for you, but can leave tasks if we have anything left to do (and I can join again a bit later)
Reminder R validation + release party is tomorrow!
https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
James is representative for all our video-off icons – staring at our code/tables trying to understand why it's behaving like it is 😜
Hackmd for listing al the problems:
*Thread Reply:* @Olivia we have put all the ones that need fixing, at the bottom you will see the ones you are working on, you can then tick them when you have solve them 🙂
*Thread Reply:* I don’t seem to have access to the file!
*Thread Reply:* you would need to log in, with you git login should work
Perhaps overlaps a bit with what the visualization team is doing, but here are some heatmaps of missingness in the three data sets (environmental, metagenome, singlegenome) ordered by publication date. The values in the parenthesis for each variable indicate percentage of missing data. Would this be useful?
Could you do the same for libraries?
sorry, James, I did not get this, missingness of what variable would you like to explore?
I was looking at the tsv-files like ancientmetagenome-environmental.tsv, and checked NA records. I guess "Unknown" should also be treated as "NA", right?
*Thread Reply:* It'll vary per column - you should check the README next to each table
*Thread Reply:* Note you should pull from dev, as Aida found an inconsistency between the two tables for archive
Making a thread of issues I’ve identified that need additional input to resolve 👇
*Thread Reply:* • ~Moguel 2021: Additional info is shown in libraries.tsv (which isn’t found elsewhere in the metadata, so there is a usefulness to having it). Example: paper shows 200_4, samples.tsv shows 200_4 , libraries.tsv and ENA show 200_4_zone3_hyposaline~
• Liang 2021: The samples.tsv only has 3/15 and the libraries.tsv has the other 12/15 (no overlap, the two together comprise the entirety of the ENA/paper entry).
• ~Borry 2020: two ID types listed in the paper - archaeological ID and laboratory ID. Samples.tsv uses labID, libraries.tsv uses archID. ENA has archID as Sample Title and labID as Sample Alias~
• Pedersen 2016: ENA sample alias and libraries.tsv file match, using style ICF40 though ENA title shows Ice-Free corridor for all. Sample.tsv shows CHL_131_11266 as sample ID, which is consistent with the paper and is also shown as the library_name in the libraries.tsv.
*Thread Reply:* • Mougel: stick with paper/samples • Liang: uhh that's confusing.. I will have to have a look • Borry2020: stick with archID
*Thread Reply:* We will need to do the same thing with the host-associated metagenomes for Borry2020
*Thread Reply:* Can you do that?
*Thread Reply:* Sure, I am almost done reviewing the env ones Aida assigned me 🙂 and I will do it after
*Thread Reply:* What's your github ID btw?
*Thread Reply:* 👍 gonna try something fancy (for em anyway)
*Thread Reply:* HEHEH i worked!
*Thread Reply:* opening pull request from command line requesting two reviewers 😎
*Thread Reply:* @aidanva @James Fellows Yates I’m thinking that the CHL IDs that are in the paper make the most sense for Pedersen 2016, does that work for y’all?
*Thread Reply:* Could you join Gather?
*Thread Reply:* Aida and I are talking about it right now in fact
@James Fellows Yates I'm free now if anything needs doing!
We will be back in about 45 minutes 🙂
*Thread Reply:* @Bjorn Bartholdy https://spaam-community.slack.com/archives/C0183TC8B0R/p1661510475199579?thread_ts=1661510101.359279&cid=C0183TC8B0R
@aidanva and myself are coming back for another hour!
Just to make sure: We prefer the "Generated FASTQ files", right? The "Submitted files" are not always available and I have seen only FTP links of the "Generated FASTQ" in the tables.
Yes always generated
I have played enough with python + ENA API for today 🙂 Looks good so far, but need to continue next week to get it productive in AMDirT!
Ardelean2020 is very confusing... will definitely need some input to resolve
Damn it, I was hoping someone would find the secret key to unlock that mess. And a nature paper at that!
I can also look this evening. Did you make any progress?
*Thread Reply:* Some. There were missing libs that I added and pushed the branch to GitHub. Not enough for a draft PR. I can update you this evening (after child-pickup and dinner)
*Thread Reply:* I'll be back about 20:30 cest
*Thread Reply:* The big problem is that the number of samples is all over the place. Supp mat has 15 (37 including replicates). samples.tsv has 16, and libraries.tsv has 30. 😫
@channel as we are getting ever closer to getting releases of the data and the tool, I would like to start gather affiliations for the AMDirT + library metadata paper!
Please fill in your details here: https://docs.google.com/spreadsheets/d/14uSdUlOpqA1rxY6j-k9M30rhSGxlFQtKxrerKudZRxU/edit?usp=sharing
For people from the first publication and who I've published with recently, I've copy and pasted some of your affiliation already but pleasereview it!
Also if you see yourself/your colleggues missing, please let me know to see why they aren't in the git logs
@Bjorn Bartholdy are you still there? I've lost all sound I think... but I can't tell
@Diana has joined the channel
1) So first thing, @channel say hi to @Diana! She is a new HiWi (student research assistant) who will be helping us with AncientMetagenomeDir, and also start extending it (e.g. looking for all exact C14 dates 😱)
2) I think we are ready to go for the big 22.09 release with all the library metadata, big thanks to everone who contributed last week! Any further inconsistencies are my fault and sorry in advance (but we can always to 'minor' releases to fix major problems 😬 )
I will do the release procedure now 🙂
Unles there is anyone interested in watching me do the release procedure itself?
(not the most exciting, but in case I'm incapacited for some reason it mighteb useful)
https://github.com/SPAAM-community/AncientMetagenomeDir/releases/tag/v22.09.2
(after a couple of mistakes from me 😉 )
@Christina Warinner stats for you in the release ntoes ☝️
thanks everyone! Next steps
- Will finish AMDirT!
- Will start planning hte publication
@channel https://twitter.com/jfy133/status/1565229326467047426
Just sent on the tweet - while I could find most people, there were a few I missed. Please PM me your twitter handle if I missed you! I've also added all your twitter handles to the manuscritp affiliation list, so please also correct me if I got it wrong there
(The main people I couldn't find, if they had Twitter or I wasn't sure was: @Arthur Kocher @Meriam van Os @valentinav and @Gabriel Yaxal Ponce Soto)
*Thread Reply:* Thank you @James Fellows Yates!
By affiliation page I mean here: https://docs.google.com/spreadsheets/d/14uSdUlOpqA1rxY6j-k9M30rhSGxlFQtKxrerKudZRxU/edit?usp=sharing
*PLEASE ADD YOUR DETAILS!*
*Thread Reply:* 2 columns "Twitter handle". Maybe you mean one of them GitHub?
*Thread Reply:* you saw nothing
Btw, @s.wasef wins on coolest department name 👀
As a reminder, what we are doing is very relevent! We are ahead of the game!
https://www.nature.com/articles/d41586-022-02820-7

So please keep an eye on our issues page and when you have a spare 20 minutes make PRs ♥️
PR for review 🙂 https://github.com/SPAAM-community/AncientMetagenomeDir/pull/938/
6 Calculus Samples / 6 Libs 👇
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/985
@channel there are loads of ancient metagenomics papers coming out! We need to make sure we keep up-to-date, please please 🙏 if you have a spare 30 minutes assign yourself and make a PR:
https://github.com/SPAAM-community/AncientMetagenomeDir/issues
@channel Ok, now conference and holiday season is pretty much over, we would like to set up our hackathon for AMDirT, in which we will need (primarily python) *developers/scripters* and *testers*.
Once AMDirT is completed and released, we will prepare the manuscript of presenting AMDirT and the library metadata update.
If you're interested in the hackathon to help us finish the tool (and learn more about it), I would like to ask which of the two days (or both) would you more likely be available for (see incoming poll):
*Thread Reply:* @Nikolay Oskolkov as you attended in the past, would you be able to join either day?
*Thread Reply:* Hi James, yes, sure, sorry for my slow response! I believe both days should work for me 🙂
*Thread Reply:* AWesome thanks 😄
*Thread Reply:* @Nikolay Oskolkov @Ian Light @Yuejiao Huang @Alex Hübner @Olivia @Shreya @Jasmin Frangenberg @Julien Fumey could you respond with a :femaletechnologist::skintone_3: emoji that you've python experience and 🐷 if you're volunteering as a tester (pig for ginea pig, which there is no emoji shockingly...)
*Thread Reply:* Holy crap @Maxime Borry you're going to have an army
@Maxime Borry will be leading the developer team, and myself or someone else will be leading the testing team
Given we have equal numbers, I'm thinking maybe we can do a two day thing even
*Thread Reply:* depending on how much work is still to be done on the tools, could be good... also would be a bit more flexible for people to be there for one day or the other if necessary
*Thread Reply:* Exactly, I wouldn't expect everyone to be there all the time
*Thread Reply:* https://github.com/SPAAM-community/AMDirT/issues
*Thread Reply:* Those are everything to do
*Thread Reply:* The main missing tooling is autofilter
What do you think @Maxime Borry?
Otherwise I'll announce it tomororw
There are 4 main things to work on: • Fix reported issues • Add requested features (mainly automated entry completion using the ENA APIs) • Testing • Writing (more) doc So maybe 2 days ain’t such a bad thing 😉
Two days it is!
Thanks for the PR @Ian Light!
Maybe someone from sedaDNA can review this? This is for Armbrecht 2022 (@Linda Armbrecht maybe, as you're in the channe land it's your data? ; ) )
But note Ian tat currently it looks like the PR comment github actions is broken, I'll need to investigate why 😞
*Thread Reply:* Ah reason why the comment is failing is because you are coming from a fork not internatally
*Thread Reply:* you're allowed to make the branch from within the main AncientMetagenomeDir repo if you want - you're a member of the organisation already
*Thread Reply:* (to make it easier on permissions stuff)
*Thread Reply:* ah OK, forgot what the Stadard procedure was and didn't want to mess anything up 😉
*Thread Reply:* You deifnitely thought right!
*Thread Reply:* But in this case it's allowed
*Thread Reply:* just don't push to master (or delete it)
*Thread Reply:* hmm will redo the PR with a branch specific thing, tried doing a quick bugfix of running the tests locally that didnt work.
*Thread Reply:* Another thing that might warrent updating is all permutations of allowed terms in the main study focus column for samples (it throws an error for Faunal,Floral but not Floral,Faunal)
*Thread Reply:* That's tricky unforutnately
*Thread Reply:* Or rather I guess it's not ,but it will make it much more filterable if we restrict to a smaller collection (as it's more consistent)
*Thread Reply:* (I also say this as I'm already not really happy about having all of these categories, because it becomes very unstanradardiseD)
*Thread Reply:* hmm maybe a discussion also for AMDirT hackathon?
Ok I'm a bit slammed today with a deadline, but I will officially announce this tomorrow on #general and via the mailing list!
But for those whove indicated above, please note down BOTH dates in your calendar!
@channel reminder we have our hackathon next week! Both (python) devs, testers (just need CLI experience), or anyone who wants to help add data are welcome!
Weds 9th-Thurs 10th November from 09:00_16:00ish CET - SPAAM gathertown
^ sooo what if your workday starts at 15:00 CET… should we show up then?
There will always be something to do :) (or I can make up things 🤣)
If anyone is in commuting distance from Germany, we will be reserving a seminar room at MPI-EVA, so you're welcome to join us!
@channel reminder for everyone joining the AMDirT Hackathon we will be starting in 35 minutes: https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
For those in Leipzig, we are in the ABI seminar room (message Maxime/I if you need picking up)
Remember that this is not just a coding hackathon, we also need testers, documenters, and will use the time to also get more of the publications in if you're not comfortable wiht coding (the normal metadata gathering you're used to to)
PLease also join <#C03BACY0CHF|amdirt-dev> as we will coordinate there from now on (as not to bother the 104 members of this channel 😬 )
@channel need some thoughts on this one on behalf of @Kadir Toykan Özdoğan.
Note anyone can give their input here, regardles if you're in the hackathon or not.
Kadir proposed adding the following paper to our environmental DNA table: https://www.sciencedirect.com/science/article/pii/S0960982221008186
Now it is shotgun data from sediment DNA. But it focuses on genomic reconstruction of human/bison/dog genomes for po(o)p-gen rather than metagenomics.
However, they did use a metagenomic tool for initial screening (Centrifuge) to identify the target species.
So question: do we consider this a metagenomic sample/paper to include in the 'Dir? Or is this too pop-genny?
*Thread Reply:* @irinavelsko @aidanva @Alex Hübner @Åshild (Ash)?
*Thread Reply:* @Pete Heintzman?
*Thread Reply:* Definitely a metagenomics sample, and would argue a metagenomics paper. As you say, there was a Centrifuge analysis in addition to the pop-gen stuff.
One could make a similar comment about pathogen metagenomics papers, where the primary focus is often phylogeography, locus presence/absence, selection, etc, rather than just a metagenomic community reconstruction. 😉
*Thread Reply:* But is it really a metagenomics when it's just centrifuge and that's it? They don't make any biological inference from the metagenomic data itself
But fair point about the pathogens...
*Thread Reply:* I agree with Pete that when the sample is metagenomic one, we should include it.
*Thread Reply:* But isn't all ancient DNA metagenomic?
*Thread Reply:* We would have to go back and add all ancient human shotgun genomes for example
*Thread Reply:* If we decide to include this one, then any shotgun genome from ancient samples should be added, not only from humans
*Thread Reply:* or do we consider anything coming from bone as not metagenomic?
*Thread Reply:* But this is primarily a sediment sample, not some petrous bone. So we expect a similar signal as for a sediment sample that was studied for microbes and not mammals.
*Thread Reply:* OK, then we include 🙂
*Thread Reply:* As it's directly from sediment
*Thread Reply:* Perhaps a definition of a metagenomics sample could be something like: “An ancient sample that is either expected or has been shown to contain genomic information from multiple taxa”
*Thread Reply:* Expected would be environmental deposits (sediments, dental calculus, etc)
*Thread Reply:* Shown would be tissue from individuals where pathogens and/or microbiomes were found
*Thread Reply:* That avoids the rabbit hole of including any and all ancient samples, which may well, but not necessarily, be metagenomic.
*Thread Reply:* by that definition, shouldn't be all the teeth samples included? we normally observe an oral signal in them, even if pathogens are not found
*Thread Reply:* Good point — do the modifications above help?
*Thread Reply:* if I understand the definition now... it has to be shown in the paper that those samples have a preserved oral signal to be included?
*Thread Reply:* That’s correct.
Is that consistent with how you folks decide on including a tooth sample in AncientMetagenomeDir?
*Thread Reply:* We would need to further refine the definition though, because a tooth may have a very minimal multi-taxa presence still. The oral signal is present (even if skewed), so we need to be a bit more precise I think
*Thread Reply:* And may need distinct diefinitions per table
*Thread Reply:* Because no, the proposed defintion does not fit here so far
> Is that consistent with how you folks decide on including a tooth sample in AncientMetagenomeDir?
*Thread Reply:* Any further comments: https://github.com/SPAAM-community/AncientMetagenomeDir/issues/1014#issuecomment-1308726818
@Åshild (Ash) we have a question for you for here: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1012
*Thread Reply:* It’s all capture data, see my reply on the pull request
any suggestions for the best way of testing changes for validation checking locally? (eg changing interior parts of validation loading, how to actually check behavior impact on the ancientMetagenomeDirCheck level
*Thread Reply:* pip install -e . in the repo while in a conda environment, but have you been working off of dev @Ian Light? It should be AMDirT
*Thread Reply:* @Maxime Borry ☝️
@Diana can you review pls: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1015/files
And @Alex Hübner @Diana for this one: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1011
For the following people, what's the likelihood you will be able to get to these PRs you've assigned yourself in the next month - we are reaching the next Dir release date in December 🙂
• @Pete Heintzman (Hebda2022) • @Anan Ibrahim (Perez2022) • @Bjorn Bartholdy (Velsko2022) • @Iseult (Hider2022) ???
Also I still need reviews for Fageräs2022 and Long2022 😞 (@aidanva @irinavelsko @Åshild (Ash) if you would have a spare 30m [lol] in the next couple of weeks
*Thread Reply:* sorry, completely forgot I did that- will get to it by the end of the week 🙂
*Thread Reply:* @James Fellows Yates I can't find anything about data availability in Hider 2022- am I missing something? It says on the issue you couldn't find it either though 😬
*Thread Reply:* Let me double check
*Thread Reply:* I don't have access to the PDF, do you ?
*Thread Reply:* (that is why I couldn't find it)
*Thread Reply:* or @Jessica Hider if you're around, would you be able to share with us a PDF of the paper?
*Thread Reply:* Ok yeah I also do dont' see anything in the main text... is there anything in the SI tables?
*Thread Reply:* If you don't see anything else then you can just leave a comment on the issue saying data isn't available and close it!
*Thread Reply:* And enjoy one less thing on your TODO list 😆
*Thread Reply:* Yeah, it doesn't seem to be there... thanks for double checking! 🙂
*Thread Reply:* That's sad though... But thanks for looking!!
*Thread Reply:* And responding quickly ❤️
*Thread Reply:* Will be done 💪 (as always shortly before the deadline…)
If any of you can't get to it please unassign yourself and we can find another volunteer 🙂
@Iseult Hi team yea sorry, not data availability yet for the paper because we are still using the data to write up a second paper and I was told not to put it up yet 😞. Data will be available on NCBI when paper 2 comes out 🙂 (which will hopefully be soon).
That's awesome to hear! Thanks for letting us know!
@channel we've been in contact with the F1000 journal and they've shown interest in publishing AMDirT there (as they have a 'special collection' of aDNA papers coming out soon). They've given us an (extended) deadline of March 31st for submission, so please expect calls for contributions/manuscript drafts being sent around in the near year
@channel reminder tomorrow 09-16:00 CET on gather town!
Documenting AMDirt and release party!
If you're not familiar with AMDirT, You can use the time to finish up your open AncientMetagrnomeDir PRs, help review the open and rest ones,, and throw in a few extra publications before I make the relesde afternoon!
*Thread Reply:* hiya james -- my morning has been thrown a bunch of random things that need to get done, but in the afternoon (1:30) I will join !
*Thread Reply:* OK no worries, see you this afternoon
I am coming in a few minutes as well 🙂
I've been sitting in the wrong gather.town la la la
@Ian Light I have a meeting at 14:00_15:00, but I can give you your tasks at 13:30 🙂
@Nikolay Oskolkov I have further tweak suggestions for you too when I'm back
@irinavelsko libs and blanks removed, thanks for catching that!
OK I have to run to Kita, but we made good progress on the docs and on covert!
I also reviewed and merged in the last few open and ready PRs and I will finish the 22.12 release this evening
Thanks to @Nikolay Oskolkov @Ian Light @Jasmin Frangenberg for joining in!
@James Fellows Yates I can finish making videos for convert and filter (I think validate is OK to just have the instructions on github) sometime on monday or early in the new year
Perfect! Yes, we only need screencast for Filter under the tutorial section so that can be a separate PR once @Maxime Borry is happy with the text docs we did today
I got there in the end, just before the holiday!
https://github.com/SPAAM-community/AncientMetagenomeDir/releases/tag/v22.12.0
Release 22.12.0 is out!
Release v22.12 includes 21 new publications, representing 187 new ancient host-associated metagenome samples, 27 new ancient microbial genomes, and 102 new ancient environmental samples. This brings the repository to a total of 153 publications, 1248 ancient host-associated metagenome samples, 560 ancient microbial genomes, and 484 ancient environmental samples
Furthermore, this release adds 323 new ancient host-associated metagenome libraries, 84 new ancient microbial genome libraries, and 123 new ancient environmental libraries. This brings the repository to a total of 2152 ancient host-associated metagenome libraries, 2180 ancient microbial genomes libraries, and 563 ancient environmental libraries.
(also I'm going to fix the 3 contributor thing now, apparently GH only automatically picks up the person who makes the release and the 'new contributors')
But I thikn I have a way around that
Ok found it, in CHANGELOGs you tag yourself so we know you've made a conribution for more visibility ❤️
And current targets for 23.03!
https://github.com/SPAAM-community/AncientMetagenomeDir/milestone/9
Thanks to @Bjorn Bartholdy @Anan Ibrahim @Ian Light @Kadir Toykan Özdoğan and @Diana!
I shall toot/tweet tomorrow!
⛄ Happy holidays everyone ⛄
Hi @James Fellows Yates I have a question about the sample_name for the Fagernas2022 entries in ancientmetagenome-hostassociated_samples.tsv
The table currently has the Pandora individual ID for these samples, but the Pandora IDs represent 4 archaeological IDs, so 4 individuals. Is it correct to have the Pandora individual IDs here, instead of the archaeological ID? There are multiple pandora IDs per archaeological individual b/c so many pieces of calculus were collected per individual
*Thread Reply:* Unlike Pandora, I don't think we are distinguishing between individuals but indeed explicit samples
*Thread Reply:* As e.g. in eDNA samples there is no 'individual'
*Thread Reply:* Ah ok. Thanks!
*Thread Reply:* It's a not as nice indeed though when you want to get to meta analyses 😕
*Thread Reply:* but I didn't want to 'hack' the other way
Hi @James Fellows Yates, The study of Farrer2021 (https://github.com/SPAAM-community/AncientMetagenomeDir/blob/master/ancientmetagen[…]ciated/libraries/ancientmetagenome-hostassociatedlibraries.tsv) is listed with the librarylayout set to PAIRED, however, but all entries have either just a single or three FastQ files (R1, R2, single). I am wondering, whether the entries that have just a single FastQ file should be set to SINGLE instead of PAIRED or was there a reasoning for setting them all to PAIRED?
*Thread Reply:* Let me have a look, IIRC this was a really wierd one
*Thread Reply:* Ah yeah, because that's what is listed on the ENA...
*Thread Reply:* https://www.ebi.ac.uk/ena/browser/view/PRJNA688065
*Thread Reply:* But agreed maybe we should fix this...
*Thread Reply:* @Sterling Wright it looks like you uploaded this, do you have any idea?
*Thread Reply:* I am not sure to be honest. I was added pretty late to the project. I will have to check with Andrew.
*Thread Reply:* I confirmed with Andrew that they are PAIRED-end. When I was instructed to upload, they only gave me those single files. Looking at it now, I am thinking some things got misplaced when Laura moved from Adelaide to Pennsylvania. I will do some investigating and see if I can find both ends of the sequences. Sorry for the confusion.
*Thread Reply:* Thank you Sterling!
*Thread Reply:* No problem. I’m sorry for this issue.
*Thread Reply:* I confirmed with Andrew that they are paired. I am in the process of tracking down each sample. It is taking a while because the data was stored at ACAD and I am trying to parse through everything here at Penn State. It may take a bit to finish it but I will upload the forward and reverse reads for each sample.
*Thread Reply:* Awesome, thank you very much for continuing to follow up!
*Thread Reply:* Thanks @Sterling Wright!
SCREENSHOT OF EXCEL yolo
@Toni de Dios Martínez do you know why ENA -converted FASTQ files aren't available for https://www.ebi.ac.uk/ena/browser/view/PRJEB49091?
@Toni de Dios Martínez has joined the channel
We are trying to add it to the Dir but can't because half the information on the FASTQ files are missing 😞
*Thread Reply:* Moltes gracies!
*Thread Reply:* Hey @Toni de Dios Martínez any update?
*Thread Reply:* yes, I have the sequences processed and already preprared to be uploaded back.
Is the interactive sample map failing to render for anyone else? Trying to use it for a presentation and this is the most I can get on either Chrome or Safari!
*Thread Reply:* I've got the same in chrome
*Thread Reply:* Me neither!
*Thread Reply:* Will try to investigate
*Thread Reply:* There are still the static maps on the main readme you can use as an interior work around
*Thread Reply:* @Maxime Borry ahy ideas?
*Thread Reply:* Thank you, and no hurry! Did a quick and dirty chart off the .tsv files which is more than enough for my needs right now :partyparrot:
*Thread Reply:* Fixed!
*Thread Reply:* Github changed their rendering URL
*Thread Reply:* Hooray! Thank you!!
@channel we are in release month again! Please assign yourself to any open issues and make a PR 🙏 , and if yo'ure already assigned to an issue please consider unassigning yourself if you don't think you can do it and post here so we can reassign!
Currently @Diana is also working on a new exciting extension (particularly for the single genome crowd - how does exact radiocarbon dates sound for your phylogenetic analyses 😉 ) so she can't cover all the main metadata PRs!
The resource is only as strong as the community ;)
Review needed on: https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1051 (enums for) https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1052
(not test for the second will fail until first is merged)
@channel for those who still have PRS to do, I will be doing a reviewing spree tomorrow, so please have your PRs open by then 😄
Ok got side tracked but reviews will definitely be doing them over the new few days
Thanks @Mohamed Sarhan @Miriam Bravo and @Kadir Toykan Özdoğan for being responsive!
Release is still scheduled for Friday(ish) but I'm now a bit sick so let's plus minus that a couple of days 😬
@channel 23.03: Rocky Necropolis of Pantalica is now out!
Release v23.03.0 includes 8 new publications, representing 110 new ancient host-associated metagenome samples, 23 new ancient microbial genomes, and 52 new ancient environmental samples. This brings the repository to a total of 161 publications, 1358 ancient host-associated metagenome samples, 583 ancient microbial genomes, and 536 ancient environmental samples
Furthermore, this release adds 180 new ancient host-associated metagenome libraries, 98 new ancient microbial genome libraries, and 64 new ancient environmental libraries. This brings the repository to a total of 2332 ancient host-associated metagenome libraries, 2278 ancient microbial genomes libraries, and 627 ancient environmental libraries.
Corrections were made to 3 publications.
See here: https://github.com/SPAAM-community/AncientMetagenomeDir/releases
Thanks to: @Alex Hübner @Mohamed Sarhan @Miriam Bravo @Kadir Toykan Özdoğan and @Diana!
Vaguely related: https://onlinelibrary.wiley.com/doi/10.1111/arcm.12869
@channel everyone who made contributions to library metadata shoudl've just recieved an email from me about the AMDirT manuscript that you are listed as a co-author on. Please read it and follow the instructions!
If you've not recieved the email but were expecting to - please DM me 🙂
@channel
> Before an article can be published, authors must suggest at least 5 reviewers who meet our reviewer criteria. Suggestion for reviewers please, in the thread below! Both PIs and postdocs are fine 🙂
Also: "Reviewers should have published at least three articles as lead author in a relevant topic, with at least one article having been published in the last five years" which is a bit shit (but it's should)
*Thread Reply:* Gabriel Renaud
*Thread Reply:* Hannes Schroeder
*Thread Reply:* Michael Knapp?
*Thread Reply:* More ideas:
Verena Schuenemann Ben Vernot Martin Sikora Michael Martin Lars Fehre-nSchmitz Laura Epp
*Thread Reply:* Ok this is really hard to find women who we aren't already cllaborating with 😅
*Thread Reply:* I don't know if their publications will be relevant enough... But from our department you could maybe check out Catherine Collins (and/or Alana Alexander).
*Thread Reply:* Kathrine Eaton?
*Thread Reply:* possibly Lara Cassidy - I know she has an interest in data/metadata management and standardisation but not sure if publications relevant enough?
*Thread Reply:* That's a very good call! Been speaking with her a few months ago 👍
*Thread Reply:* Ludovic Orlando, Tom Gilbert, Felix Key?
*Thread Reply:* The first two are too high up to consider IMO, the last is @Ian Light supervisor 😬
*Thread Reply:* Thanks though! Keep the mcoming!
*Thread Reply:* Tina suggest: Mikkel Winther-Pedersen
*Thread Reply:* Maybe final list:
Mike Bunce, Katherine Eaton Lara Cassidy Mikkel Winter-Pedesen Mike Martin Hannes Schroeder
Hello @channel!
Everyone has now responded confirming authorship and the waves number of comments has now reduced.
I will submit the preprint today and make the reviewer suggestions today - but if you've not yet given your comments - please don't worry as we are still in the preprint stage and can make tweaks/suggestions!
Thank all for your help and work on the short time frame ❤️
Will confirm once the submission is made 🙂
*Thread Reply:* Sorry, I haven’t had time to make my comments, but will try to do asap
*Thread Reply:* No worries! Can come during preprint period like I said 😄
Everything uploaded, including copying once again all 40+ affiliations and 20+ funding information.
Maxime and I are just writing teh cover letter (because I totally forgot)
Final reviewer suggestions:
Communciated with the editor some instituae overlap but I think it should be OK
Amazing! Thank you @James Fellows Yates and @Maxime Borry
@channel!!!
I'll make the fixes this afternoon/evening if I have time!
Remember the system is in reverse for F1000 you get 'published' without review (just copy editing) and then this is somewhat akin to a pre-print but already citeable etc and with copy editing. The reviewers will then review, and then based on those comments we make a new version. This new version will then have ticks next to it showing it has been peer reviewed and no different from a traditional article
@channel anyone around for a very rapid QC check on a slightly revised abstract?
Basically we need to convert it to a BAckground/M<ethods/Results/Conclusions structure
I basically split up the original into Background/Methods/Conclusions and added a results section, thoughts?
*Thread Reply:* I would include the first three lines of the Results in the Methods too.
*Thread Reply:* Three sentences or lines?
*Thread Reply:* Isn't the 5000 ancient metagenomic libraries results?
*Thread Reply:* ```\textbf{Background:} Access to sample-level metadata is important when selecting public metagenomic sequencing datasets for reuse in new biological analyses. The Standards, Precautions, and Advances in Ancient Metagenomics community (SPAAM, \href{https://spaam-community.github.io}{https://spaam-community.github.io}) has previously published AncientMetagenomeDir, a collection of curated and standardised sample metadata tables for metagenomic and microbial gene datasets generated from ancient samples. However, while sample-level information is useful for identifying relevant samples for inclusion in new projects, Next Generation Sequencing (NGS) library construction and sequencing metadata are also essential for appropriately reprocessing ancient metagenomic data. Currently, recovering information for downloading and preparing such data is difficult when laboratory and bioinformatic metadata is heterogeneously recorded in prose-based publications.
\textbf{Methods:} We present an extension of AncientMetagenomeDir that provides standardised library-level metadata of ancient metagenomic samples, and the companion tool `AMDirT', which facilitates automated metadata curation and data validation, as well as rapid data filtering and downloading.
\textbf{Results:} Through a series of community-based hackathon events, AncientMetagenomeDir was extended to include standardised metadata over over 5000 ancient metagenomic libraries. The companion tool `AMDiRT' provides both GUI and CLI based access to such metadata for users from a wide range of computational backgrounds. We also report on errors with metadata reporting that appear to commonly occur during data upload and provide suggestions on how to improve the quality of data sharing by the community.
\textbf{Conclusions:} Together, both standardised metadata and tooling will help towards easier incorporation and reuse of public ancient metagenomic datasets into future analyses.```
*Thread Reply:* That's the code if you want to just change it and show me waht you mean 🙂
*Thread Reply:* ok I see, yes the extension to 5000 libraries is part of the results. But I see the part of how we made it to 5000 libraries as methods (e.g. “Through a series of community-based hackathon events”). but I may be wrong.
*Thread Reply:* So move community-based hackathon events to section above :+!
Thanks @Iseult! Can I have one more?
```Dear James
Facilitating accessible, rapid, and appropriate processing of ancient metagenomic data with AMDirT Borry M, Forsythe A, Andrades Valtueña A, Hübner A, Ibrahim A, Quagliariello A, White AE, Kocher A et al.
We have now accepted your article for publication in F1000Research. It will be sent to the typesetters and a member of the Production team will send you a proof in due course.
Please note that your article will only be published once you have suggested 5 suitable reviewers who meet our reviewer criteria. Please do not contact your suggested reviewers, as this has the potential to influence and invalidate their review. Our editorial team will contact any suitable reviewers on your behalf and will be your main contact once your article has been published.
Best wishes,
Cecilia The Editorial Team, F1000Research ```
@channel AMDirT co-authors the proofs are ready!
I'll make any changes and submit on Friday morning (CEST)
Feel free to look through and double check, and report any problems to me! **DO NOT MAKE ANY CHANGES YOURSELVES!
And as always, don't panic if you miss the deadline, we will release a new version once the reviews are in, and can incorporate changes then :)
The proof of your article F1000Res134798 is now available for review at:
The proofing system is web-based and allows you to directly incorporate your corrections into the electronic files. The interface to make corrections works in a similar way to Word with track changes. A guide to help you use the system is available at the following link:
https://ops.spi-global.com/eProofingF1000/ViewProofingHelpGuide_Journals
N.B. Please use Google Chrome or Mozilla Firefox browsers. These are the current browsers that support the system.
Please follow the instructions given in the proofing system regarding corrections and complete your review within 3 day(s) of receipt of this email.
You are responsible for correcting your proofs. Errors not found may appear in the published journal. The proof is sent to you for correction of typographical errors only. Revision of the substance of the text is not permitted, unless discussed with the editor of the journal. Any necessary changes should be entered via the proofing system. Please be sure to answer all queries present, as the system will require you to resolve the queries before submission of corrections. Any outstanding queries are listed in a column to the right of the proofs.
If you are unable to meet this deadline, if you encounter any problems, or if you have further questions, please e-mail the below address and reference your article ID in all correspondence.
Please note that connecting an ORCID account to F1000Research requires the account holder to sign in to both F1000Research and ORCID, therefore it isn't possible for us to add ORCID badges for your co-authors on their behalf. When the article is published, they will receive an email encouraging them to connect their ORCID account to F1000Research. If they do this, their ORCID badge will be displayed next to their name.
If there are any outstanding queries on your reviewer suggestions, then we will be in touch with you shortly.
Best regards, Michael Production Editor
production.research@f1000.com
71 proof changes and 22 comments 🤦 but it's away! lets see if they come back again
Dear James
Facilitating accessible, rapid, and appropriate processing of ancient metagenomic data with AMDirT
Borry M, Forsythe A, Andrades Valtueña A, Hübner A, Ibrahim A, Quagliariello A, White AE, Kocher A et al.
Please click here to download the PDF proof of your F1000Research article.
Please look through the article and let me know if it requires any corrections or if you are happy for it to be published as it is. Please also confirm the following details are correct:
All author names are spelled correctly
Authors are listed in the correct order
Affiliations for all authors are accurate
The information in the Copyright section is correct
All figures and figure legends are correct
All external files, including data files are correct
All links within the article are working, and correct
Please note that connecting an ORCID account to F1000Research requires the account holder to sign in to both F1000Research and ORCID, therefore it isn't possible for us to add ORCID badges for your co-authors on their behalf. When the article is published, they will receive an email encouraging them to connect their ORCID account to F1000Research. If they do this, their ORCID badge will be displayed next to their name.
Corrections at this stage may require further typesetting and therefore cause some delays. If any corrections are necessary, please mark them directly on the PDF file using the commenting and markup tools in software such as Adobe Reader.
Please return your proof corrections to us via email - please note that after the article has been published, any requests for minor corrections will only be considered on a case-by-case basis. Therefore, we encourage you to check your proofs carefully at this stage.
If there are any outstanding queries on your reviewer suggestions, then we will be in touch with you shortly.
Best regards,
Jessica
The Editorial Team, F1000Researc
If anyone has any time to look through it this evenig!
Note it does seem correct that citations come AFTER full stops/punctiation (makes no sense to me but OK)
*Thread Reply:* My affiliation isn't correct anymore (it was when I looked at the first version). My name should have 16, Anatomy Department, University of Otago, but my name says 27. And @Ian Light's name has 16 now instead. 🤔
*Thread Reply:* My affiliations are also incorrect now - they should be 38 and 39 rather than 17 and 18 🙂
*Thread Reply:* Kevin's is also incorrect and should be 38 not 17
*Thread Reply:* Also my affiliation (and Nihan's) is wrong. 21 rather than 32. Thanks James
OK, I've just looked through and it looks like the proofing people COMPLETELY messed up teh affillations
I'm very qnnoyed, they've ignored a lot of other comments too apparently
Probably won't submit here again (which is sad as up until proofing, this was pretty good experience)
Please look very closely at the rest of the manuscript everyone!
Lets find all the other stuff they've messed up 🙄
Ok apparently they correctly addressed all the main text commetnts and affiliations but ignore everything in the 'header' page...'
OK I've replied saying do not publish until the corrections have been made!
*Thread Reply:* Hi James, found these typos: Page 3 line 3: 'AMDiRT' should be 'AMDirT’ Page 5 line 2: line should end with a point, not comma. Page 10 line 14: ‘greater or equals’ should be ‘greater than or equals’ Page 11 line 38: ‘headers of the’ is redundant. Page 13 line 5: ‘ancient DNA’ should be aDNA
*Thread Reply:* Thank you very much - 🦅👀!
I'll see if they will accept these in the next round of the proof once they've fixed the stupid affiliations
*Thread Reply:* (I made have [professionally] rage replied )
*Thread Reply:* Hi @James Fellows Yates, I read the proofs (quit carefully) and did not notice any major bugs except for the affiliations 😞 you guys did a very good job with the text! If I were to pick on small details:
- page 7, in the sentence "To install, users are recommended to use the pip or Bioconda package managers", I would make "pip" and perhaps "Bioconda" italic, or use quotes to highlight it somehow, as it is a tool / terminology and (I suppose) not a regular English word 🙂
- page 9, figure 2: the images seem too small to me, I can barely see the menu items that the cursor points to, but I understand that it is hard to make the size and quality much better here.
- page 12: in the sentence "For example, mapped BAM files include only reads mapped to a particular reference genome and thus do not represent a full metagenomic dataset.", I am not sure that this is true. I guess it depends on the aligner. At least, if no samtools filtering with respect to MAPQ is done afterwards, I am pretty sure that Bowtie2 and BWA (as well as e.g. STAR, just on the top of my head) include all reads, i.e. mapped and unmapped, into their BAM-alignments. So in theory, unfiltered BAM should be equivalent to raw fastq. But in general I agree with the message that fastq should absolutely be preferred for data sharing compared to BAM 🙂
*Thread Reply:* Hi Nikolay,
I'll try and incorporate the changes if we get another chance
- Ok, sounds easy enough
- The HTML version (which is the main one for F1000 as it's not a Print journal) can go full size so you will be able to see :)
- You are right, I think I even mention the uBAM format, but I didn't want to get into too much detail, mainly to avoid people thinking it's alright when the fastq should be standard and what most metagenomic tools accept
*Thread Reply:* @Nikolay Oskolkov I've just looked through the proofs again
Re 1. I realized we've been not done that for any other tools or libaries throughtout the text, so I'm reluctant to go and modify all of them now (as technically we shouldn't be making any changes other than major ones...).
I think it's not too bad as in the next 5 lines we show specific commands from the tools, so I think even if it's unfamiliar with a reader, they should be able to infer what they are.
That said, reading that section did make me notice we had a separate mistake where we called it a 'Bioconda package manager', where bioconda is the repository not the manager itself, so I will correct that with the updates from Nihan.
*Thread Reply:* Sounds good @James Fellows Yates! Also if not too late I would replace "ameta" in the references with "aMeta" as this is how we typically write it 🙂
*Thread Reply:* It already it is?
*Thread Reply:* Oh in hte references
*Thread Reply:* That's odd... I pulled it via cross-ref...
*Thread Reply:* Hi James sorry, I am looking at this now. I hope everything is fixed bc I saw our affiliation is also messed up.
Still waiting on the corrected proofs (got a very nice apology from the proofing editor, with detailed explanations though!), however in the meantime the work doesn't stop here!
We have lots of issues to be integrated before the next release end of this month!!!
Please have a look at the publications list here: https://github.com/SPAAM-community/AncientMetagenomeDir/issues
And assign yourself to make a PR, but also please keep an eye on the Pull Requests tab to help out reviewing 🙏 . Note though: please only assign yourself to an issue when you're definitely going to work on it and/or have already started. This will help @Diana (who's back from thesis submission/excavating) prioritize which issues she should do :)
Deadline for this reelase: June 30th!
I will try and work on getting autofill working ASAP to help the procedure along
@channel
Ok attempt FOUR for the AMDirT manuscript!
I've been communicating with a very nice and responsive production editor now (so better experience now ;)), and apparently we've found a few bugs in their production platform that actually affected other manuscripts too 👀.
Here is the latest version, where apparently the correct affiliations , should be associated with the right person. However for those with multiple affiliations: apparently their system does not allow for the order of the affiliations within each person's to go in the order we originally requested, as it forces alphabetical order. So please note that that will be not be possible but given its clearly alphabetical I hope that is not a problem. If it is please provide a workaround by re-wording your affiliation.
Aida and I are now travelling for the next 3 days, so I won't be able to look myself until probably Monday, so please take the chance that your affilTions are correct in the meantime!!
*Thread Reply:* Alas, my name has superscript 41, which corresponds to the wrong institute.
*Thread Reply:* @James Fellows Yates my affiliations should be 22, 23 (order does not matter) and not 33, 34
*Thread Reply:*
*Thread Reply:* Yes, mine should be 30! Unless I’ve unknowingly been hired by the Pasteur… c’est pas mal!
*Thread Reply:* Yes, I'm afraid mine are still wrong and should now be 38 and 39 🥲
*Thread Reply:* Yeah, mine are wrong too. They should be 2, 4 31, and 32
*Thread Reply:* Mine and Valentina's are also wrong 😶 Should be 21
*Thread Reply:* i hate to add to the fire, but I am with 16 which should be 37 :')
*Thread Reply:* Mine is correct! :allthethings:
*Thread Reply:* Mine is right as well 🙂 just to give a good news :)
*Thread Reply:* For me the first is correct (2) and the second should be 33. The way it is now in the paper is also how is was before. I am not sure if it’s the same for others, but for my affiliations there were no corrections done, basically.
*Thread Reply:* In my case I have 13 while it should be 34
I just got another version, cold someone check if this one looks right? Train WiFi is being 💩
https://f1000research.com/articles/147881/pdf
*Thread Reply:* …sorry James, I am now from Trento, Italy
*Thread Reply:* @James Fellows Yates nothing seems to have changed for my wrong affiliation 🙂
*Thread Reply:* I have the same number 41 as before, UChicago has the same number 30 as before, but my number 41 now corresponds to a different institution from before
*Thread Reply:* The easiest would be just to manually correct the affiliations, there are not so many co-authors
*Thread Reply:* I have no control over this ;(
*Thread Reply:* I am still 33,34 while I should be 23,43
*Thread Reply:* Maybe I am doing something stupid but I get this
*Thread Reply:* I logged in with my google account credentials
*Thread Reply:* I did try with those hmmm weird
*Thread Reply:* I have the same issue as @Maria Lopopolo
*Thread Reply:* Uhh ok @Nikolay Oskolkov could you download the PDF and share it here?
*Thread Reply:* Or @Shreya if she's around!
*Thread Reply:* The new version in question ^
*Thread Reply:* I'm sorry you have to go through this James.. But since the affiliations are in a different order, both of mine are wrong now.
*Thread Reply:* Do we have an original list of people and affiliations somewhere? Perhaps to send to the editors so they can at least do a preliminary check for correctness before sending us another version?
*Thread Reply:* Looks like @Gabriel Yaxal Ponce Soto @Julien Fumey and I, have also wrong affiliation (13 instead of 2 for Yaxal and I, and 13 and 20 instead of 2,9 for Julien).
*Thread Reply:* Sorry James this is a pain!
*Thread Reply:* @James Fellows Yates Even yours and mine are now messed up 😂
*Thread Reply:* Yes I'm look at it now
*Thread Reply:* and saw the same thing
*Thread Reply:* For me: 5 became 16, still 5 is written next to my name. 🫠 Fun fun fun, sorry
OK SO I think they have a bug in their system.
Doing a bit of proof-hsitory archaeology (hehehehe) It seems their system deals with affiliations in blocks of 11. And for whatever reason, each proof seems to shuffle these blocks around in each version
Soooo lets see how the next attempt goes (just emailed them)
Greetings from a random service station in the middle of Valencia!
Please check again, I've had a quick skim and I think it's ok now?
*Thread Reply:* Correct! Thanks for taking care of it!
*Thread Reply:* Correct 👍 Just missing ORCID (didn't notice it before, but don't mind if it's too late for that now). https://orcid.org/0009-0004-5961-4709
*Thread Reply:* Orcids come from your f1000 account
*Thread Reply:* Not from the submission
*Thread Reply:* Make sure you use the same email on f1000 as with your orcid
*Thread Reply:* Thanks, will create one.
*Thread Reply:* Mines are correct too! Thank you !
*Thread Reply:* I confirm mines are also correct 🙌
" The affiliations – we are honestly just as confused by this whole thing as you are! We really do apologise for the continued errors – I wish I had a better explanation for you other than we’re still investigating and trying our best to fix. There seems to be something happening in the download of the pdf, both via author accessible proof links and our own production editor buttons. We see the affiliations correctly assigned (albeit in a slightly different order to your corrections), but when the proof is downloaded they seem to randomly assign themselves into a different order and that’s what you see when you download. This is a lovely new addition to the error we’re experiencing across articles where pdf affiliations aren’t matching the system and web articles, and the development team are aware and attempting to fix.
We can download the pdf from our own system in a couple of different ways, and one of those ways should yield the affiliations that match (attachment). We’ve checked these against your list of correct affiliations and the assignments should be correct, but the alphabetical ordering seems to have just decided not to work – potentially due to some attempted fixes by our development team. Could you check the attached that the actual assignments of the authors are correct (and if not please indicate which ones are incorrect)? We’re also interested to know how important the actual ordering of the affiliations is per author i.e. when an authors is assigned to 2 affils. Are you happy for any list order so long as they appear at least in order of appearance in the list, and assigned entirely correctly?
We definitely won’t be putting this article live with this mis-matching issue still happening, and will ensure we have your approval of the affiliation list before we do.
Thanks for your continued understanding and patience, and apologies again"
Btw: we are waiting for bug-fixes to their backend system but it seems the production editor knows now what the afiliations should look like to tell the technical team if the fixes are working. We should receive one final proof once they are done and it'll go live
*Thread Reply:* I got a reply thsi evening sadly not:
*Thread Reply:* ```Hi James,
Thanks for the message. Unfortunately, a permanent solution hasn’t been found and the issue is still affecting live articles. We apologise for the delay, and we’ll be in contact when its fixed with your full proof to check before we publish.
Many thanks,
Jess```
*Thread Reply:* Are they saying that we identified a bug that's affecting all their articles?
*Thread Reply:* WE DID INDEED 😄
*Thread Reply:* Quite fundamental actually
*Thread Reply:* ```Hi James,
Thanks for the response – we appreciate you finding the time to check the article.
The grant info has now been corrected on our system and will pull through correctly to the final version.
The affiliations – we are honestly just as confused by this whole thing as you are! We really do apologise for the continued errors – I wish I had a better explanation for you other than we’re still investigating and trying our best to fix. There seems to be something happening in the download of the pdf, both via author accessible proof links and our own production editor buttons. We see the affiliations correctly assigned (albeit in a slightly different order to your corrections), but when the proof is downloaded they seem to randomly assign themselves into a different order and that’s what you see when you download. This is a lovely new addition to the error we’re experiencing across articles where pdf affiliations aren’t matching the system and web articles, and the development team are aware and attempting to fix.
We can download the pdf from our own system in a couple of different ways, and one of those ways should yield the affiliations that match (attachment). We’ve checked these against your list of correct affiliations and the assignments should be correct, but the alphabetical ordering seems to have just decided not to work – potentially due to some attempted fixes by our development team. Could you check the attached that the actual assignments of the authors are correct (and if not please indicate which ones are incorrect)? We’re also interested to know how important the actual ordering of the affiliations is per author i.e. when an authors is assigned to 2 affils. Are you happy for any list order so long as they appear at least in order of appearance in the list, and assigned entirely correctly?
We definitely won’t be putting this article live with this mis-matching issue still happening, and will ensure we have your approval of the affiliation list before we do.
Thanks for your continued understanding and patience, and apologies again,
Jess```
*Thread Reply:* And earlier message from her
*Thread Reply:* We just claim a bug bounty compensation, maybe we can ask for no APC 😂
*Thread Reply:* Likely it's a subcontractor so I doubt that will go through
Good morning everyone!
We have a release imminent at the end of this month (next week), it looks like all but one of the proposed papers have been assigned, so it would be great if we can have help reviewing (this is just checking for accuracy of the proposed metadata!), please let me know if you would like more guidance on this!
Anyone available to review a 1 sample 2 library PR?
https://github.com/SPAAM-community/AncientMetagenomeDir/pull/1088
Good morning all. Does anyone have a few extra minutes today to review Clavel 2023, please? It's a long one, though!
*Thread Reply:* I'll be on it after lunch!
*Thread Reply:* But if anyone else is willing to tke over and get it done today they will be my favourite person of teh day
FYI last update was from 5th of July:
I’ve nudged our development team again but unfortunately they say that since a fix hasn’t been found yet, they can’t project when it’ll be pushed to our system. I know this isn’t ideal at all, but you’re at the top of our list as soon as we know more.
On August 5th I'll give them a deadline for August 19th (which will be 3 months since initial submission()
@channel please check for accuracy of the numbers with your affilation - note that the order per person may not be excact (e.g. FSU comes before EVA for Maxime), but each number affilation is with the right person now I think?!
*Thread Reply:* At this point, it's also fine with me this way 😉
*Thread Reply:* Yes agreed 🤦♀️:skintone3:
*Thread Reply:* Affiliations 13 and 20 are correct 😀
Please confirm, and when a few people have said it looks good to them I'll let the type setter know!
*Thread Reply:* Mine is good too 😊
*Thread Reply:* All good, just I my orcid is not shown (it is linked to f1000)
*Thread Reply:* If you linked it after we submitte dit won't come up until later
(also please congratulate the new Dr. @Maxime Borry who passed his PhD defence today! 👏👏👏)
Telling the typersetter it's ready, so I expect the 'pre-peer review' to be out within a week
```Dear James,
I’m pleased to let you know that your article: "Facilitating accessible, rapid, and appropriate processing of ancient metagenomic data with AMDirT" has just been published on F1000Research.
The Peer Review Process
We are now inviting the reviewers you have suggested. As part of their reports, reviewers are asked to provide a recommendation of 'Approved', 'Approved with Reservations' or 'Not Approved', and their report will be published alongside the article with their full name and affiliation. You will be able to respond to any published reports with a comment and/or by publishing revisions as a new version of your article – we will send you instructions on how to proceed when you begin to receive reports.
It is important that authors do not contact the reviewers directly, as this could result in invalidating their report.
Please note that we will ask you for additional reviewer suggestions if the invited reviewers decline. In order to avoid any delays with the peer review of your article, please continue to check your Suggest Reviewers page for updates and respond to any email requests as soon as you can.
Linking Your Data
Now that your article has been published and assigned a DOI (10.12688/f1000research.134798.1) we would strongly recommend that you include this DOI in the metadata of any published dataset associated with this article. If you would like assistance with this, please contact our editorial team.
Increasing Discoverability
Now that your article has been published, why not take advantage of the tools we provide to help maximize your article's reach and share your article using the Email and Share options on the article page.
- Add your article to your ORCID record (or create an iD here) to ensure you receive full recognition for your professional activities.
Your article will be listed in ePMC shortly as a preprint, and then updated once it has passed peer review.
Kind regards
The F1000Research Team
Press releasing articles: Please avoid promoting articles in the media until the article has passed the open peer review process. Promotion on social media is encouraged once the article has been published; please ensure the full citation is included, as this contains the peer review status. F1000Research should be cited as the source of these articles with a link to the article.```
Congratulations @channel!!!!
*Thread Reply:* @Kadir Toykan Özdoğan @Maria Lopopolo maybe schedule tweet/toot for next week?
*Thread Reply:* (preprint though, it's not peer-reviewed!)
*Thread Reply:* Press releasing articles: Please avoid promoting articles in the media until the article has passed the open peer review process.
*Thread Reply:* Read the next sentence
*Thread Reply:* So we are published (social media allowed), not peer-reviewed (no newspapers)
*Thread Reply:* @Kadir Toykan Özdoğan @Maria Lopopolo probably time to post!
*Thread Reply:* Ok I will post it!
@channel due to 'bad coordination' Diana is on sick leave and I'm trapped underneath a baby, so it would be great to have some community help again keeping the 'Dir ticking along.
We currently have 5 unassigned issues open, from all three tables so it would be great if we could have some people addressing them! Most are quite small :)
We have a release deadline end of September which is why I bring it up, so you should have plenty of time
(X3 pathogen, X1 single dental calculus, x1 environmental study)
*Thread Reply:* I got the environmental one
Would this be valid?
https://link.springer.com/article/10.1007/s13313-023-00936-6
*Thread Reply:* Looks reasonable: shotgun libraries with SRA raw data available.
@channel it would be great if we could get some more reviewer suggestions, F1000 are finding it difficult...
*Thread Reply:* has someone already suggested anders bergstrom? think he did a preprint a while ago about data availability & aDNA
*Thread Reply:* Technically I was acknowledged on that preprint 😬... butttt not a coauthor... I guess we could try? I don't know who he may have published with recently
*Thread Reply:* Ah @Ophélie Lebrasseur has published with him 😕
*Thread Reply:* And @Pete Heintzman
*Thread Reply:* Unless we can make some argument about it
*Thread Reply:* (not metagenomics, and on large mega papers...)
*Thread Reply:* Xuexue Liu published software for visualising ancient sample metadata if that qualifies
*Thread Reply:* Oooh yes, link?
*Thread Reply:* https://academic.oup.com/bioinformatics/article/38/16/3992/6623404 for the paper
*Thread Reply:* Ooooh yes perfect!
*Thread Reply:* You could maybe try Logan Kistler for broad ancient genomics / appreciation of large datasets, he might show mercy
*Thread Reply:* Fair I don't think anyone in the AMDirt paper worked on plants much recently...
This is where we are currently
It would be nice to have people in any (but not all!) of the areas: ancient DNA, metagenomics, and/or computational/metadata stuff
@channel reminder next release is in 9 days, it would be great to have the PRs ASAP to give time for reviewers to do their business :)
Also I'm sad to announce that @Diana will unfortunately be leaving the AncientMetagenomeDir team at the end of the month.
She's done a great job at keeping us up to date, so let's give her a round of applause👏👏👏👏l
This also means the community will need to again take up the slack (heh). If there are any newcomers who don't know where to start, please let me know and we can arrange some training for you
Once I'm back from parental leave I will also introduce the next publication plan for the Dir :)
Thanks for the kind words, @James Fellows Yates , I really had a great time being a part of this team and I am sad to go! Just a quick note - I will be here until the end of the next month, so let me know everyone, if I can help with anything in the meantime.
@Diana @Cameron Ferguson @Kadir Toykan Özdoğan @Anan Ibrahim @Iseult release is end of this week if you can get the last couple of PRs open)
*Thread Reply:* will do my best, but realised after the fact that there wasn't any new data in the issue I assigned myself (from this: https://www.nature.com/articles/s41467-023-41174-0#Abs1 ) - it looks like some would have previously been uploaded as part of other studies, but others are from human aDNA studies, so might not already be there. should this be uploaded at all? If so, should I just do the libraries that haven't been put into the dir, or is there a way to add data for previously published samples with a different species?
*Thread Reply:* I left a comment on the issue to Diana actually - they are new genomes so we should still list them! We just point to the original accession codes
*Thread Reply:* A previous example is Lugli https://github.com/SPAAM-community/AncientMetagenomeDir/issues/1109#issuecomment-1725871842
*Thread Reply:* So basically add as normal: add the new genomes to the single genome table and the corresponding libraries just re-using the ERS/SRS codes/lib metadata from earlier studies if we already in clude them 🙂
*Thread Reply:* For human aDNA studies, then it'll require new ERS/SRS codes and gathering those lib. metadata info
*Thread Reply:* Please let me know if that doesn't make sense, I'm running on 4h sleep atm,
*Thread Reply:* ok I’ll do my best just been quite busy over the last few days 🙂
*Thread Reply:* @Cameron Ferguson I can take over yours, if that helps, but I don’t see which ones you’ve signed yourself up for 🤷♀️:skintone4:
*Thread Reply:* @Iseult - Same applies to the Hodgkins 2023 issue - I can take over, if you like. 🙂
*Thread Reply:* That would be super helpful if you've time @Diana - I can do the review if that would speed things up as well? 🙂 Thanks!
*Thread Reply:* @James Fellows Yates Ok that should be all info added now, last remaining item is updating the change log. what information should I included for that?
*Thread Reply:* Project name, DOI and your GitHub tag. Look at the othersfor examples
*Thread Reply:* would I put it under added or changed seen as its updating an existing entry with missing samples?
*Thread Reply:* Ah good point, no in this case just add your name to after Dianaes
*Thread Reply:* As it's inclusion is new to this release
*Thread Reply:* should be good to go
If anyone could make an issue (and/or even better a pull request) for the following that would be awesome (dealing with a baby with a cold and 2 days with only 4h sleep 😭 and now the GitHub app keeps breaking my phone [!?!!?!?])
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-023-01647-2

*Thread Reply:* Thanks @Alex Hübner!
(Museum) Frog gut microbiome?!s
https://github.com/SPAAM-community/AncientMetagenomeDir/issues/1116
Would people feel more motivated for helping our reviewing if we also listed reviewers of each PR on the changelog (as public evidence of your contribution)?.
If yes 👍 on this message
Calling out for help on mastodon for reviewers for AMDirT might have worked! Have a professor(!) In the US who said he is happy and willing to review!
Ok, sorry @Diana @Iseult given there are still open comments on Hodgkins and it's a complicated one lets bump that to the next release (I need to do 23.09 today). Please keep at the update/review loop until you're both happy and get it in ASAP so we odn't have to think about it again 😆
And it'll be a nice big one for 23.12
WE HAVE OUR FIRST REVIEW!
*Thread Reply:* As in accepted by F1000, or as in approved by the reviewer ?
*Thread Reply:* Approved by the first set of reviewers :)
*Thread Reply:* We have at least review incoming
*Thread Reply:* ok. I guess we should wait for the other review(s) to start answering then 🙂
*Thread Reply:* But most of the comments are straight forward.
The only two more involved things (outside of rephrasing) is adding some form of library of metadata filtering and maybe a little community event to test on as many machines as possible installation of the tool
*Thread Reply:* The former I have at least a UI PR open
This paper describes the ‘AMDirT' open source software for interaction with ancient metagenome sequence sample metadata tables. The tables have been produced through a massive communal curation effort by the international SPAAM (The Standards, Precautions, and Advances in Ancient Metagenomics community) group to systematically improve on what is available from the public repositories of archived short read sequence data. AMDirT allows viewing and searching of the tables using the Python streamlit library and facilitates download of data and integration with open source nextflow analysis pipelines. AMDirT also helps with initial creation and validation of new metadata tables.
I successfully installed AMDirT version 1.4.6 using conda on my Intel MacBook following instructions in the manuscript. I was able to replicate the commands described. There is also a public server <https://www.spaam-community.org/AMDirT/> for the AMDirT viewer. The documentation for the software is very good and includes video tutorials.
The manuscript is well-written and clearly outlines the functions of the software. The development of AMDirT represents a significant effort, not least because of the community-wide consultation. The need for community-based metadata addresses a well-known problem with the SRA/ENA/DDBJ databases and the discussion provides some nice examples of the type of issues faced when downloading. AMDirT is a valuable tool for both the ancient metagenomics research community and those outside the community interested in browsing and accessing the data.
Specific Points
The streamlit-based viewer is pretty slick but I feel the importance of command line interface (CLI) is a little underplayed here. The viewer can be used by people without CLI proficiency but to actually take actions like making new sample tables, or download and process data, CLI is essential.
The instructions in the text and on github for conda install should guide users to install into a fresh conda environment rather than into the base.
There should be a list of computational environments that the software has been tested on (e.g do M1-3 macs work?)
I did not see a mention in the text that there is actually a public facing server <https://www.spaam-community.org/AMDirT/>
“Newly added library information columns include the library name (how data are typically reported in original publications), the aDNA library generation method (e.g., double-stranded or single-stranded libraries), the library indexing polymerase (e.g., proof-reading or non-proofreading), and the library pretreatment method (e.g., non-Uracil-DNA Glycosylase (UDG), full-UDG, or half-UDG treatments). The latter three fields represent information about the sequencing library construction that influence the presence of aDNA damage, a factor that is critical for the processing of aDNA NGS data.8,18 Sequencing metadata columns include instrument model, library layout (single- or paired-end), library strategy (whole genome sequencing, targeted capture, etc.), and read count. “ I could not work out how to search these fields through the viewer. These are accessible as downloads post validation but it seems that users would want to search through samples based on these fields?
Further questions
Finally, I have some three questions about the project that it would be great to get comments on.
How sustainable is the community effort to maintain these databases moving forward into the future with the probability of the number of samples increasing each year?
Have you tried sending the improved metadata back to SRA/ENA?
How difficult would it be to adapt the software for a different community of researchers that wanted to improve annotation but use fields that would be different from the ancient metagenomes community?
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Ye
Second AMDirT review!!! Not approved right away, but comments mostly positive and more about reworking the text to satisfy his curiosity (as far as i can see). Seems to have a bug too.
It seems we have one more incoming, then we can plan how to address everything
https://f1000research.com/articles/12-926/v1#referee-response-210768

I found a mistake: The coordinates for sample SK8, “St. Mary Magdalene leprosarium, Winchester”, Schuenemann 2013, are wrong. Lat should be 51.059 like Sk14 (Mendum 2014), not 52.059.
Oops, could you make an issue?
<#C0628MAM54H|ancientmetagenomedir-c14-extension> 👈
> If I'd like to invite someone to give PAASTA a little intro about how to make community-curated version-tracked citable resources (like AncientMetagenomeDir) through GitHub, would you be willing to give such an intro talk? Or, if you're too busy, could you recommend someone I should contact? @channel cause we are clearly the best and everyone wants to copy us, is there anyone interested in presenting how AncientMetagenomeDir works to the new palaeoproteomics community?
*Thread Reply:* Is there a possible date for that seminar?
*Thread Reply:* In December
*Thread Reply:* Is what I was told
*Thread Reply:* Ok will message you with more details
Hello all, I have a quick question. I am a bit confused by the F1000 setup. I assume from the previous messages that the AMDirT article is not officially published and still under review?
*Thread Reply:* Yes, we have two reviews (they are public, you can see them on the article page), but I don't have enough time to work on it at the moment
*Thread Reply:* That's no problem at all! I just wanted to check the paper status for my MSCA report. Thank you!
*Thread Reply:* Can consider it 'under revision'
*Thread Reply:* One review approved, the other tentatively approved with (minor) comments really
*Thread Reply:* If you want a more 'traditional' category
Hi @channel, I hope you are all slowly getting in the holiday mood with the end of the year slowly approaching. This also brings along another release deadline for AMDir. At the moment it's relative quiet there. There are in total five low hanging fruits. The majority of issues have already been tackled by@Diana and I will do the reviews while @James Fellows Yates is on parental leave. However, there is another issue about a sedaDNA study by Perez et al. (2023; DOI: https://doi.org/10.1111/fwb.14182) that would still need some volunteer? Any sedaDNA person that has time to tackle this publication?
*Thread Reply:* Hi Alex, I got the Perez et al. paper!
Another thing: while I try to do my best to sit-in for @James Fellows Yates while he's gone, I am not as good as he is in spotting publications that are relevant for AMDir and should be added. If you spot one that's missing, please follow the example of @Kadir Toykan Özdoğan and others and just open an issue yourself here: https://github.com/SPAAM-community/AncientMetagenomeDir/issues If you are not familiar, how this works, just contact me! I am happy to help! Cheers!
Can someone add the following to the issues?
*Thread Reply:* And @Abby Gancz @Sterling Wright can you already guess the two things I'm going to ask 😅😬
*Thread Reply:* I'll create issue in a moment.
*Thread Reply:* Hey @James Fellows Yates yep I'll get to it as soon as I can. Next week okay?
*Thread Reply:* Also uploading the unmerged/untrimmed reads?
*Thread Reply:* That might take a little longer but it sounds like Laura is good with us doing that
*Thread Reply:* That would be wonderful thank you to all :) really helps with FAIRness of the dats 💪
Hi @channel
We've noticed a marked drop off in the amount of contributors/submissions over the year, and I would like to better understand why.
I've made a very short(! - max 2-3 minutes of your time) anonymous survey where you could give feedback. There is one multiple choice question, and one free text question. Neither are mandatory so you can pick either or.
Any and as much feedback as possible will be gratefully received to help make your lives easier and maintain the project over a longer period.
You can get to the anonymous form here: https://form.jotform.com/233351873343052
And please be as honest as possible (emphasis on anonymous again :))
@channel reminder of the short survey above! Thanks to the 9 responses so far! Getting about 20 would be perfect: the short anonymous survey is for both new and existing contributiors
Hi channel, Happy New Year to everyone! As previously mentioned I would like to prepare the next official release of AMDir in the next days. There are still a few open issues and PRs so I just wanted to quickly check in what the status is here:
• Perez2023: @Kadir Toykan Özdoğan ? • Gancz2023: @Abby Gancz (see my comments on your PR)? • Murchie2023: @Kadir Toykan Özdoğan? For White2021, were you able to upload the sequencing data once more to ENA, @Toni de Dios Martínez?
For Caruana2023, while there has been some movements and the authors finally seemed to release the project (https://www.ebi.ac.uk/ena/browser/view/PRJNA934104), there are still no sequences uploaded nor have the authors responded to any email we sent. So I would hold these back.
If I could get feedback on the other four open publications this week, I would make a call, which of these will be included in the next release. I would like to publish the next release next week.
Another to add: https://www.nature.com/articles/s41598-023-48762-6
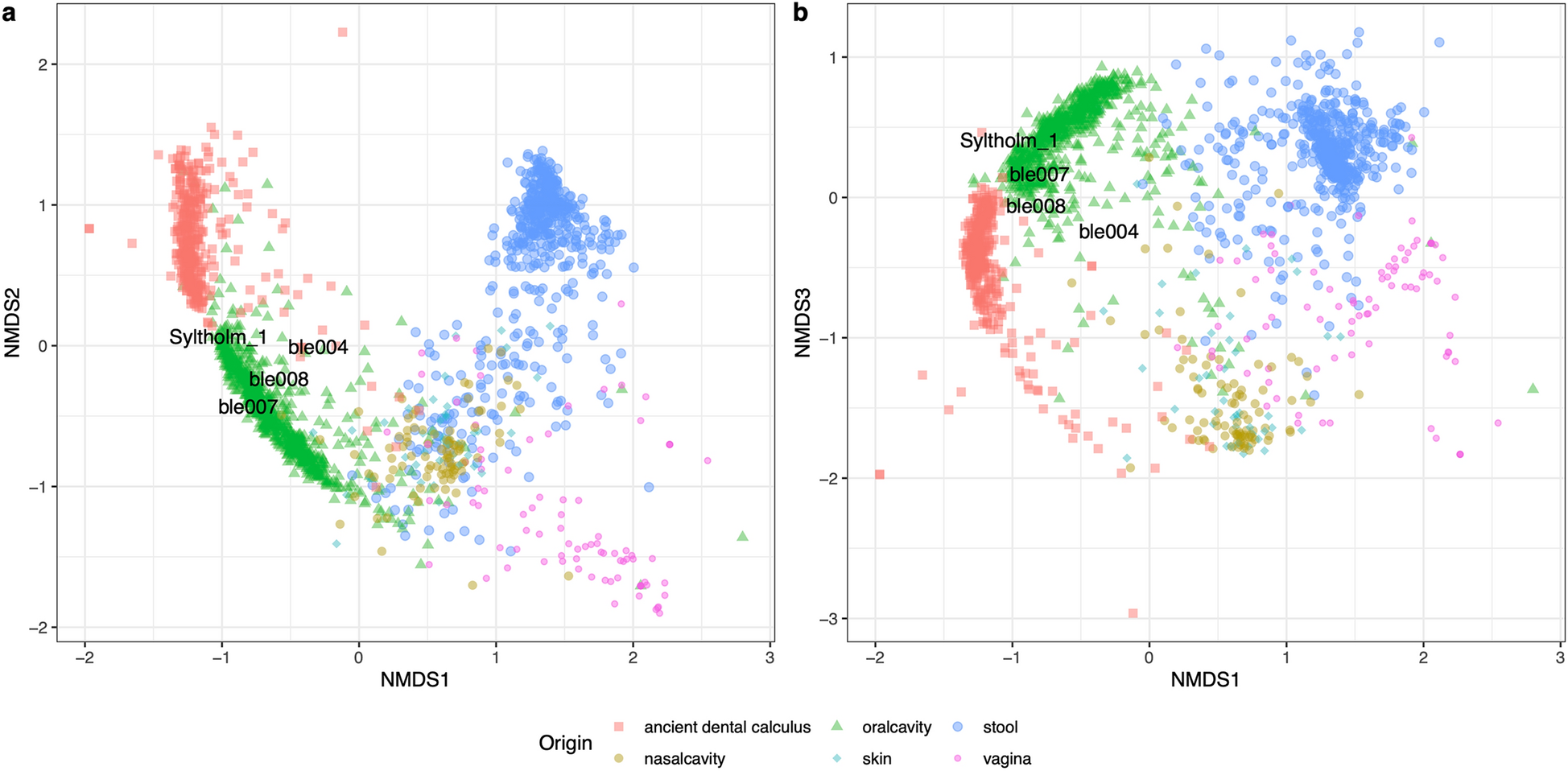
https://www.nature.com/articles/s41586-023-06965-x
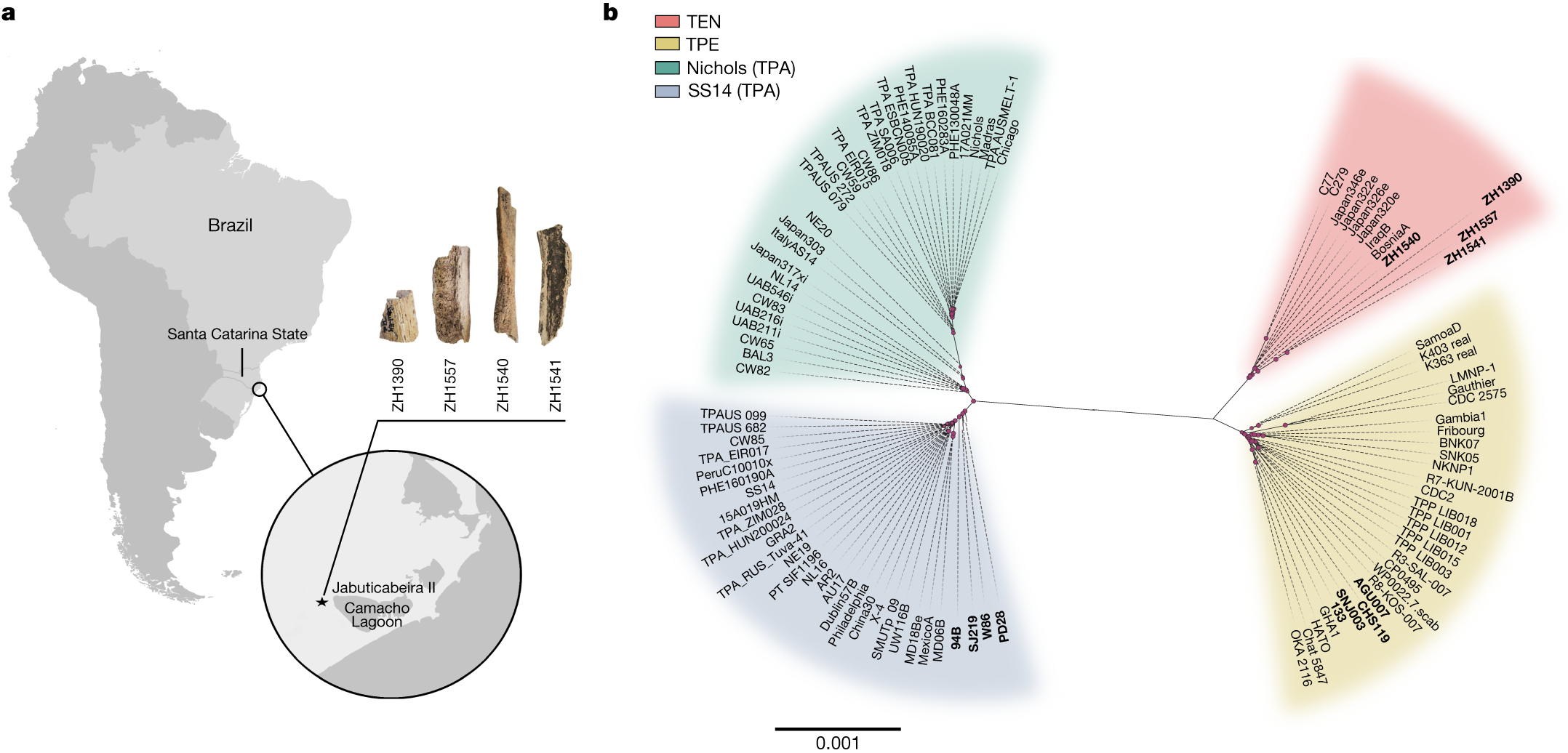
*Thread Reply:* i can grab this one sometime this week 🙂
Pathogen genomes if anyone is interested
(just one by the looks of it so should be pretty easy)
Palaeofaeces if anyone is interested
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0295924
Hi @channel,
This is the announcment of the release v23.12 Cathedral, Alcázar and Archivo de Indias today. Release v23.12.0 includes 3 new publications, representing 11 new ancient host-associated metagenome samples, 38 new ancient microbial genomes, and 30 new ancient environmental samples.
Thank you everyone who put in the work to make this release possible! ❤️
We are currently a bit out of sync regarding when we preferentially want to submit the next release and when we did it. However, currently, there are a number of new publications in the issues section and I think it would be great if we would manage to have these ready by the end of March.
As James mentioned a while ago, there is a drop in the participation of the project (https://spaam-community.slack.com/archives/C0183TC8B0R/p1701505117017479). Some of you filled out the form and gave us feedback why this is the case. Based on your feedback, James and I have come to the conclusion that we should have a open discussion about the next steps of AncientMetagenomeDir. We will send out another message to schedule such a meeting in the next weeks.
Note at least 3 of the publications for March are single genome/pathogen studies, so should be quite straight forward 🙂
@channel we have a release scheduled at the end of this week, but we've only had 2 PRs merged in and 7 publications/issues not yes addressed - any chance we could have a few volunteers?
*Thread Reply:* @Anan Ibrahim @Kadir Toykan Özdoğan you both are assigned to one each, do you think you can still make it by end of the week?
*Thread Reply:* yup, will be done by tom. 👀
*Thread Reply:* Yes 👍:skintone2:
Also whoever volunteers can be beta tester for cool new functionality to (hopefully) make your lives much easier 😏 , so please let me know if you do and I can give you instructions 😄
Four sample calculus PR from @Marica Baldoni @Claudio Ottoni if anyone is is free for a quick review 🙂
@Anna White Is it correct yuo have two PRs open for the same publication?
*Thread Reply:* Yes, sorry, I made a mistake but I’ll delete one.
*Thread Reply:* Ah ok, then I wait 🙂
*Thread Reply:* It should be correct now! But please let me know if anything needs changing 🙂
*Thread Reply:* Ok will do! You're missing a CHANGELOG file update
*Thread Reply:* But otherwise:
- I' m going to try something quickly, please ignore the next two comments
- I will aim to review tomorrow :()
*Thread Reply:* Thanks! I managed to make a bit of a mess of it but I’ve added the changelog update and it should be good now 🙃
*Thread Reply:* Looks ready for review 😄
*Thread Reply:* @Anna White how did you get the exact lat:lon?
*Thread Reply:* From their first publication on these samples, they have the exact location in the supplementary files.
*Thread Reply:* The pop-gen one?
*Thread Reply:* Such a weird way of doing it with those maps...
*Thread Reply:* But ok thank yo 😄 it otherwise looked correct, was just surprised how specific it was 😬
*Thread Reply:* (that's unusaual)
*Thread Reply:* Haha, yes, it was a bit of a search but that was the only mention of a specific location I could find.
@channel hope you had a good long weekend if you/your country partakes!
Reminder we have a release scheduled *end of this week*, so please open/finish your PRs, I will do a reviewing blitz on Friday and do the release then (maybe with @Alex Hübner if he's free/around). We still have 3 issues/publications not yet assigned (two pathogens, one metagenome)!
Also please see the post from Alex on #general https://spaam-community.slack.com/archives/CPYE64ZC6/p1711540972353019 and fill in the when2meet attendance will be important for discussing the future of the project!
OK last ping @channel, but mainly because we have very few existing contributors signing up 😞 ☝️
(consider this is a light guilt trip, you aren't obligated of course 🙂 )
@Iseult @Piotr Rozwalak I've reviewed both of your PRs! They aren't far off 😄
I will aim to do the release midday tomorrow (instead of morning when I normally do it), to hopefully give you both a chance to get into the next realse 🙂
@Anan Ibrahim I think Alex gave you comments already
@channel AMDirT F1000 paper co-authors: please check your emails! Maxime and I just sent an updated manuscript based on reviewer comments for re-submission 🙂
You have ~3 weeks to send comments and we attempt to resubmit
And otherwise I'm going to do release 24.03 now 🙂 thanks to all the last minute PRers!,
Oh and also: new AMDirT command that @Maxime Borry wrote in 1h 😎:AMDirT, you can now chain download -> convert entirely on the CLI if you're a poweruiser 🙂
In case you've muted #general 😏
@channel future of ancient metagenome dir meeting zoom link is now open!
TL;DR @channel Alex is the new project lead 👏 👏 👏 So please welcome him. He plans to set up a dedicated core team and more regular trainings and events, and help facilitate projects around AncientMetagenomeDir. If you're interested please let him know - he will set up another meeting for setting up the core team. The core team can consist of veterans and complete newcomers 🙂 so don't be shy
Hi @channel, As James mentioned in his message last week, we have decided that it would make a lot of sense if we would have a dedicated core team for AncientMetagenomeDir that helps to keep the project running instead of burdening it all on one person's shoulders.
We had in mind that the major responsibilities of the core team would be: • provide teaching material how to use the AMDir for research • be available for open-office hours to answer questions related to AMDir • provide teaching material how to add new data to AMDir • organise hackathons to add new data to AMDir • brainstorm and discuss potential additions of new data types to AMDir • brainstrom and discuss new community projects to AMDir I hope it's clear from this list that we look for a diverse group of individuals. It doesn't really matter at what stage of your research career you are, whether you are a AMDir novice or expert, a superb programmer or a communications expert. In the end we will need all this different types of expertises to make AMDir work and I hope you will learn skills from your peers that you might not currently have.
Therefore, I would kindly ask the individuals who would like to be involved in the AMDir core team to fill out their availability for an inaugural meeting in the following survey: https://www.when2meet.com/?24693334-Zs4X7
In case you have further questions or aren't available at any of these dates but still want to be involved in the core team, please comment on this thread or message me directly.
I am very much looking forward working with all of you!
Hi @channel, Thanks to those who participated in the survey. Based on the votes, I would suggest to schedule the AMDir core team meeting for Wednesday, the 8th of May, at 3 pm CEST. I will share a Zoom link here for the meeting in the next days.
There won't be a formal agenda for this first meeting other than to discuss how we should set up and run the AMDir core team and what our initial steps should be. If you can spare a moment, you can maybe think about what would be important for you?
Have a great weekend and see you next week!
https://spaam-community.slack.com/archives/C03BACY0CHF/p1714987640567319
Here is the Zoom meeting link for our inaugural AMDir core team meeting:
Topic: AMDir Core Team meeting Time: May 8, 2024 03:00 PM Amsterdam, Berlin, Rome, Stockholm, Vienna
Join Zoom Meeting https://gwdg.zoom.us/j/85278050066?pwd=UTluSFdCRXhva2RwMjg5c3NnRU55dz09
Meeting ID: 852 7805 0066 Passcode: 052805
See you all later!
Hi @Diana, @Bjorn Bartholdy and @Emily Gaul were are about to start the core team meeting.
*Thread Reply:* I sincerely apologise to everyone for missing the meeting just now.
*Thread Reply:* Sorry from my part as well! I was away on holiday 8th and 9th (which were tentative plans that I forgot to add to my calendar, so I'm also sorry if I indicated that I was available!)
Typeset revision of AMDirT/AncientMetagenomeDir2 paper!
*Thread Reply:* Here are my comments @James Fellows Yates
*Thread Reply:* Thank you @Maxime Borry! Will add to the list, I also found the wierd broken link thing
*Thread Reply:* I don't htink I'll ask them to put the 'provided here' link though...
*Thread Reply:* have you seen it? 😅
*Thread Reply:* https://github.com/10XGenomics/supernova/blob/b16b613b14687efe20a6ad33161cb461dae04699/tenkit/lib/python/tenkit/illumina_instrument.py#L12-L45
*Thread Reply:* Changes sent!
Please have a look through and see if you see any mistakes/typos etc @channel!
I will re-submit corrections tomorrow, so you have until then 🙂
V2 is apparently live! (A bit unexpected)
https://f1000research.com/articles/12-926/v2
🤞 That we bump the second review to complete approval and then we can say task well done 👏








































 }
}

 }
}



 }
}










 }
}


















 }
}