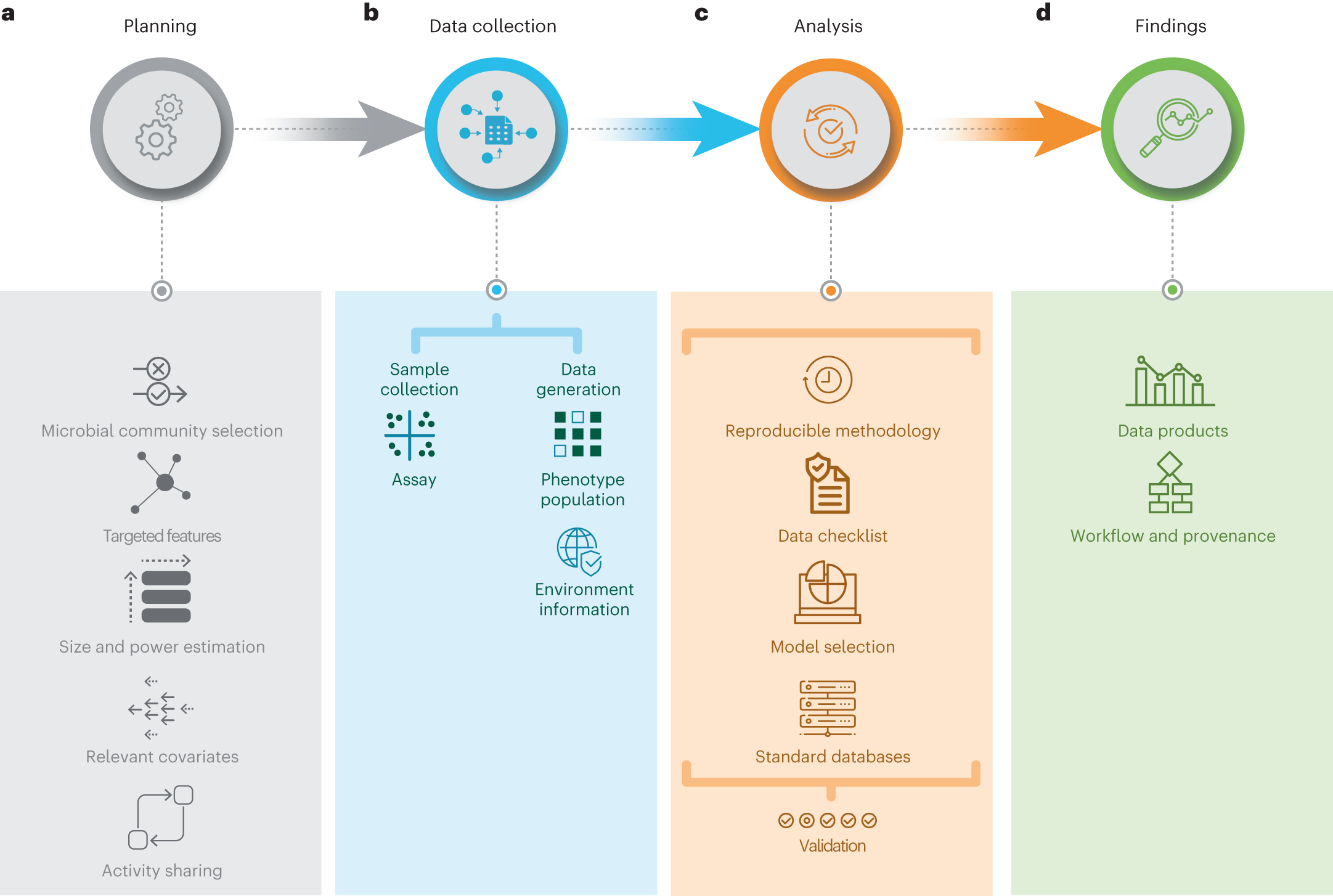
Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
@Antonio Fernandez-Guerra has joined the channel
@Ophélie Lebrasseur has joined the channel
@Clio Der Sarkissian has joined the channel
@Katerina Guschanski has joined the channel
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008260
@Gunnar Neumann has joined the channel
Just came across this: https://microbiomedata.org/
I wonder if an arch-sci standardised metadata format would fit that 🤔
@Laura Carrillo Olivas has joined the channel
I've contacted the ENA accordingly:
```I saw the announcement about the plan to require collection date and geographic location of the samples of sequenced data uploaded to the ENA.
1. I was wondering if there is already some guidance or specifications already existing publically that can be viewed already in regards to this, i.e., would this just be enforcing existing metadata specifications, or expanding upon them?
2. I ask as I work in a department that generates a lot of ancient DNA data, and I am a member of a community that is aiming to standardise a lot of metadata reporting for ancient metagenomics. We have found that although the palaeogenomics field is extremely good at uploading all data to places like the ENA, the quality of metadata uploaded in these contexts is quite poor. This is partly as many of the fields do not really work for ancient samples.
In particular the 'collection date' field could be an issue for 'ancient' remains. For example, for museum samples - the 'collection' date could refer to:
- The age of the sample itself from when it was living?
- e.g. for archaeological material, often via radiocarbon dating, and can reach back tens or hundreds of thousands of years
- The date of deposition in the museum or collection?
- as recorded in museum collections, however for older collections this may not exist
- The date the sample for DNA extraction was actually taken (e.g. powder drilled from a bone?)
3. I would also be curious to know if the ENA would be interested in supporting extension to current metadata uploads sheets to add additional fields specifically for ancient samples (e.g. an ancient extension of MIxS or MIMS sheets). An initial step would be this would primarily be supporting 'age of death' type of dates, e.g. radiocarbon dates etc.
I ask as there is a lot of interest both in the ancient metagenomics community, and in palaeogenomics in general. However there are not so many experts in regards to data upload etc.```
*Thread Reply:* Please keep me on the loop
*Thread Reply:* Was planning to already 😉
*Thread Reply:* Given 'the plan' 😬
Please let me know if you want to know the repsonse (@Antonio Fernandez-Guerra maybe you would be interested in being kept in the loop)
We got a reply!
Hi James,
Thank you for your email.
1) We are likely to re-use existing metadata structures for this (e.g. the
existing fields 'collection date' and 'geographic location (country and/or
sea)' in samples) but specific details are still under discussion as we
receive feedback from users. We will be releasing more details by April 2022.
2) Thank you for your comments, we recognise that ancient DNA is an area we
don't yet fully cater for so it would be useful to know how we can better
capture/support submission of ancient samples. In general, 'collection date'
is described as the 'date of sampling' and the 'date that the specimen was
collected' so I would generally advise for this to refer to the sample
collection event (e.g. the date of the find). It is possible that we could
provide additional standard fields, for example, to capture the estimated age
of an ancient sample, which would be optional for specific use cases like
this.
3) Yes, we would be interested in this - if you are able to share a list of
minimal metadata to capture for ancient DNA samples (like a starting
checklist) we can look into this for you. Additionally, if you have an
interest in developing an official MIxS extension for ancient DNA samples, I
could put you in touch with the Genomic Standards Consortium.
Best wishes,
Josi
I reply saying essentially yes to everything 😬
@James Fellows Yates keep me in the loop if they put you in contact with Lynn Schriml or Chris Hunter from GSC
Then we can continue from where we left it last year
and you can take the lead
I've also passed the info on to the human poop-gen peeps so we have the side too. I'll keep everyone informed here as well
Thanks @James Fellows Yates for pushing this 💪
Doodle poll for the ifrst meting!
I would basically go through the procedure of how this is apparently meant to work, and ideally if people could look through existing checklists we could already start comparing notes 👍
@channel first meeting will be January 28th 09:00_10:00 CET!
Google calendar event is available https://calendar.google.com/event?action=TEMPLATE&tmeid=MTIzZnByNHVsaTVpY3BlMjk5c2FjM3RxamsgamZ5MTMzQG0&tmsrc=jfy133%40gmail.com|here
We will meet in the SPAAM gather.town
For people who can't make that time - don't worry, you can still be in the working group, I will send around meeting notes as well, and you can still post here feedback already (I aim to send instructions today).
I will aim to optimise meeting times in the future so more people can meet (unfortunately this is a little restricted in January for various reasons on my end)
More meetingh details here: https://spaam-community.github.io/#/events/minas-working-group-jan2021/README?id=metadata-standards-working-group-meting-jan-2022
In particular please see the Prequisities section to know how to prepare!
@channel for all the those who are joining the MInAS checklist working group, I would ask if you could already go through the attached spreadsheet and already review the metadat fields already there.
Please indicate in teh yellow 'minas' column of the MIxS_V6 tab whether you think the corresponding field would be relevant for ancient data. Column descriptions are available halfway down here here: https://gensc.org/mixs/ (i.e. what the M/C/X/E/- specifications mean). You can ignore the environmental tab for now.
Please send me back your verison of the file before the meeting (with your initials in the name), and I will aggregate them together
If oyu want to read the original two MIxS papers, PDFs are here:
https://paperpile.com/shared/8hysxH https://paperpile.com/shared/qvmm47
And a more recent one for MAGs: https://paperpile.com/shared/jLe6ra
@Kendra Sirak has joined the channel
Updated sheet to have a comment colunn (@Kelly Blevins I'll update your one for you)
Also - if you're torn between two of the 'strictness' values (e.g. mandatory vs. conditional mandatory), then just opt for the looser one!
@channel reminder we have the first meeting THIS FRIDAY at 09:00 CET on gather.town
Please go through the spreadsheet above, but also @Antonio Fernandez-Guerra and I have prepared a draft project proposal to submit to the GSC. Please leave comments with any feedback! The whole thing is up for discussion
https://docs.google.com/document/d/1XiKRcP_3nVvp914RnF2Lyxsw5iLZNQirQP1d275GtCM/edit?usp=sharing
I went through the doc and made some comments, focusing on trying to equilibrate between ancient DNA from remains and from environmental archives (not easy though). I think this is very good! Looking forward for progress :)
*Thread Reply:* Perfect thank you!
*Thread Reply:* Looks great, James — suggestions and comments have been made.
*Thread Reply:* Awesome hanks!
@channel reminder of our meeting TOMORROW at 09:00 CET, please if possible have gone through the file as described above and send it to me before the end of TODAY
https://spaam-community.slack.com/archives/C01BX7EM4EL/p1641829141003200
(I need a bit time to aggregate it- htanks for the 4 who have already done so!)
I'm in gather.town, ping me here if you have any problems connecting!
Shared Meeting notes (everyone can add stuff!): https://docs.google.com/document/d/1WO7gFU9TcEGzJEt5exW-LqCC_4gw7eMVxSzI5Knrp0I/edit?usp=sharing
Where can I find the gather.town link @James Fellows Yates?
I first uploaded to the SRA in 2014 and it was traumatic! 😆 None of the fields made sense and there were no instructions. We can do better!
Traumatic for me too (last year, did not recover yet 😅)
But if you can believe it, uploading proteomics data to the PRIDE repository is worse
https://spaam-community.slack.com/archives/C01BX7EM4EL/p1642145829005600
*Thread Reply:* For sedaDNA we can ask Viv, Mikkel and Linda to join the subWG too
*Thread Reply:* @James Fellows Yates I will be in microbiome and sedaDNA
*Thread Reply:* @James Fellows Yates, could I also join the microbiome WG? Thanks 🙂
*Thread Reply:* Yes of course!
@channel meeting notes from todays meeting! Feel free to add anything I missed, or if anything is unclear! https://docs.google.com/document/d/1WO7gFU9TcEGzJEt5exW-LqCC_4gw7eMVxSzI5Knrp0I/edit?usp=sharing
Doodle poll for the next whole-SPAAM MINAS meeting: https://doodle.com/poll/9iinimpav8ecbqni?utm_source=poll&utm_medium=link
Also the name MInAS was something I chose as a temporary thing, so if you have a better acronym for the project don’t be shy
And @Meriam Guellil your definition of conditional mandatory was basically correct: https://github.com/GenomicsStandardsConsortium/mixs-rdf/issues/24
Oh lord, if you go to the github repo to find the latest version, you get sent in circles 😆
Everything is inactive and sends you to the legacy one which says the latest one is here but this one is inactive
@channel PLease remember to start filling your your availability for the next whole-SPAAM meeting here! https://doodle.com/poll/9iinimpav8ecbqni?utm_source=poll&utm_medium=link
You should already have been added to sub-working group specific channels, and hopefully started coorindating there (well done to team pathogens who look like they've already started!)
@channel doodle is now shut! The next whole-SPAAM MINAS metadatastandards meeting will be on *March 4th 2022 at 14:00_15:00 CET * on gather.town!
Google calendar invite thingy (and/or send me your google accounts if I don't already have it) https://calendar.google.com/event?action=TEMPLATE&tmeid=ODd2YjN2bmk2bDBlOXVlODJoZXBraTRzczggamZ5MTMzQG0&tmsrc=jfy133%40gmail.com
Please make sure you have your sub-working-group meetings and have your draft MIxS checklists (+ env packages) ready by then!
@channel reminder that we have the next MINAS metadata standards meeting THIS FRIDAY at 14:00!
We will be reviewing the sub-discipline specific sheets we should have generated over the last month, and we can identify overlaps or places where we agree or disagree, and discuss definitions etc.
@Anan Ibrahim @Kelly Blevins can you please share your final conensus sheets here so everyone can familiarise themselves already
Here are our thoughts from the pathogen side: https://docs.google.com/spreadsheets/d/1m8aW6EWc19ASvqIkNpSnKPvXKW-_ZWr1qqjessvz2lc/edit#gid=1290739534
Overall, our biggest concerns/points of discussion for the next meeting are
- How do we deal with taxon/organism-specific fields when the raw data from one library may contain >1 studied microbe from the publication and will contain many unstudied microbes not reported/analyzed in the publication?
- Is it necessary to repeat content between base ENA/SRA upload, MIxS core, and EnvPackage fields? For example, can we just include the project name in the base ENA metadata entry, or do we have to repeat it for each package?
- Do we want to include metadata that is independent of the sample and the uploaded raw data? For example, do we want to include fields for screening and decontamination software and parameters?
- Finding the balance between conditional and optional: will we be able to create drop down menus or yes/no questions on which metadata reporting can be conditional?
Here is also a copy of the sedimentary ancient DNA group comments regarding the core ancient DNA requirements and sedimentary ancient DNA in particular: https://docs.google.com/spreadsheets/d/1WYgLzzWItCbrKK1yRlhdoI2ng0veU_FaYUUG06iwsyg/edit?usp=sharing
@channel don't forget we will meet in the SPAAM gather.town!
https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
https://docs.google.com/spreadsheets/d/1qFKaltoadMEVM8ILY171rtdkhHUM6KkEVul_ug3fzlg/edit?usp=sharing ⬅️ all the consensus tables combined
@aidanva @irinavelsko can you hear us?
@channel for those who joined on Friday: thanks again for attending (and sorry that it was a bit inconclusive, I didn't plan ahead enough after kita closures).
I've gone through the 3 checklists and consolidated them properly this time:
https://docs.google.com/spreadsheets/d/1qFKaltoadMEVM8ILY171rtdkhHUM6KkEVul_ug3fzlg/edit?usp=sharing
While we wait for the GSC, I would be grateful if you could send me comments on the above. In particular, check the MINAS final consensus column on MiX Core Consolidated, and see if you think if you have further suggestiosn to resolve anything lsited as 'CONFLICT' (it may just be everyone agrees, just had a slightly different concept of mandatoriness, so if you think it's OK to change that to reach a consensus then let me know).
Furthermore, I've gone through MInAS specific, and consolidated these as far as I could . I've indicated lines in yellow when I believe a term already exists in MIxS. These ones will be our biggest contribution to MIxS/GCS so we will talk about these in depth in the future. Please check if you agree we can drop a term (if one already exists), or if I've 'broken' anytihng/something doesn't make sense
(Note most of my queries on MInAS specific is for sedaDNA: @Anan Ibrahim @Pete Heintzman)
Just a small update, due to a GSC conference happeinng this week(?), the GSC commitee has been very busy. However they responded yesterday and a meeting has been set up in April.
Will keep you all informed as we hear back!
OK @channel update from @Antonio Fernandez-Guerra had a good meeting with the GSC, who explained the new workflow they have developed.
What we need to do is go through the fields that the different groups disagreed on, and clean up half-written descriptions to make a more polished first draft of the checklist.
Please keep your eyes peeled for after May for a new meeting scheduling for this meeting!
Right, @channel I'm one day late (sorry!)
Lets see up a meeting. I would like to set up a longer meeting this time (maybe a whole afternoon/morning, if possible).
I think what I would like to do is for everyone who attends is that we would all split up into discipline-mixed groups of 2-4 people, then each group would be assigned a bunch of terms based on the ones we identified would be relevent for aDNA. Then each group would polish up the definition/descriptions etc, and decide precisely on whether it's mandatory/conditional etc. We would then finalise based on the new latest release of MIXS and the nwe can formally apply to have the corresponding files on the main MIXS checklist
The thing holding us back is that we have open questions/half written descriptions
Please fill in the doodle pll within the next TWO WEEKS: https://doodle.com/meeting/participate/id/aQWZXDGd
@channel reminder about the doodle above ☝️ had very few responses so far 😞
PLease let me know if July is awkward becaus eof holidays or something
@channel last reminder of the doodle above! I will be shutting it tonight 20:00 Berlin time!
This will be an important one because we will be working on polishing our proposals for each field that we will be presenting to the GSC
@channel next meeting for MINAS is July 14th, 13:00_16:00 (Berlin time) https://gather.town/app/PlXjb0deog0B4JCq/spaam-community
Before the meeting, please review everything in the attached excel sheet that is included under the MINAS column (i.e. anything NOT -)
Remember you can still join if you did not sign up on the Doodle!
@channel reminder meeting is next week ☝️ !
Meeting is tomorrow! Please make sure you've reviewed the MINAS columns in the excel sheet above!
I also would like to discuss ideas how to try and get input from the wider aDNA community, so please think if ideas (surveys, conferences to present at etc)
And we will be working on here: https://docs.google.com/spreadsheets/d/15AoMk3xNTK6XsoLQQ6M3e6wd2ZF8SuaAy87XMey4bvk/edit?usp=sharing
Starting in next couple of minutes @channel!
Can meet in the ocnference room thing with the whiteboard 🙂
Here is the v6 changelog https://github.com/GenomicsStandardsConsortium/mixs/blob/main/changelogs/v6-change-log.tsv
@James Fellows Yates we cannot write
@James Fellows Yates Can you come talk to me and Arthur for a second?
@James Fellows Yates For the reviewed part, should be reviewed by someone who is not the one modifying it?
@Antonio Fernandez-Guerra No, review just mean completed
If your group has finished, feel free to take a 5 minute break!
Then we can meet abckat 14:30 CEST and talk about the dissemination question (if you still have time)
I can do 10 more minutes
To summarise the outcome (but read the notes for more detial):
• We went through and did a clean up/polishing pass of our current draft • I will complete this to try and clean up the remaining questions over the next 3 months (I'll contact specific people as necessary) • Once this is done we will meet with the GSC ◦ This is also important as there is some structural problems that we have to know how different fields/checklists link each other • Antonio will begin on a website • We create a timeline for the next steps ofthe projects (how to get people from other fields involved, and how to 'sell' to diferenet people)
👋 everyone, sorry for the radio-silence again. Here comes some updates!
I finally had time to polish up the draft we went through last time. You can see this here:
https://docs.google.com/spreadsheets/d/15AoMk3xNTK6XsoLQQ6M3e6wd2ZF8SuaAy87XMey4bvk/edit?usp=sharing
(note I've changed the permissions for reasons below, please send me your google account if you wanted to by added for editing rights)
We now have a website - http://mixs-minas.org/ (note this is in beta, so might be buggy, let me know if you have issues)
With these two, @Antonio Fernandez-Guerra and I will formally approach the GSC to get approval of the project, and then we will start making github issues on the GSC website where we can start discussing the terms.
So once we have that OK, we will set up our next meeting to talk about how to disseminating our draft for feedback and getting more input!
Also I will add a contributors page, and list everyone who has been in the meetings (according to the meeting notes), so please check if I missed anything (will do it in asecond)
Hi @channel very sorry for the lack of updates, Antonio and I met again as the current coorindators to discuss 2023
Summary:
• Antonio to email GSC on monday to get official go • We will both look into setting up in-person/hybrid 'regional' meetings where we invite different aDNA groups from different research areas to go through the propsoal for feedback (this will require funds we expect)
Next update - GSC meetings have been pushed back so waiting for that to be rescheduled
In the meantime, I've prepared an abstract to present at ISBA (where we may try and arrange a discussion session there to get feedback already):
https://docs.google.com/document/d/1nKIPADCBAfltn0rybQtZnrjPKjIg4LJ4oyqAQZafIdY/edit?usp=sharing
*Thread Reply:* I like! Added minor grammar comments
Also, is there anyone who would be interested in being a per-country/region representative? (e.g. I was thinking @Kelly Blevins for UK if she's still based there)
*Thread Reply:* I’m still here! And happy to rep the UK. Would this involve circulating the checklist with UK aDNA labs and collating feedback?
*Thread Reply:* I can't say we've got a good mechanism yet of doing it though
*Thread Reply:* So would be something for us to discuss
*Thread Reply:* Alternatively we could try applying for a workshop grant I'm the UK to pull people to you instead?
*Thread Reply:* What sort of funding options are there for a workshop grant? I am potentially supportive of a workshop, but I am also aware of everyone’s time. But at least there is some accountability and incentive to engage if there is an actual event to attend.
*Thread Reply:* I think I will also engage archaeologist and bioarchaeologists more broadly to get feedback on the archaeo metadata
*Thread Reply:* > Funding options > Very good question... @Antonio Fernandez-Guerra and I are looking for national ones currently (but not much success for DE ATM). Other ideas I've had suggested to me is Wenner-Gren maybe? Or for UK what about Wellcome trust?
*Thread Reply:* > Supportive of a workshop > It's up to you ultimately as well how to execute the best way but we need to maximise participation
*Thread Reply:* > Engage archaeologists > Yes that would also be ideal but I feel we need a more polished draft already first
*Thread Reply:* I think our lab would be interested, especially @Abby Gancz. I was thinking maybe a workshop at SAAs or ABBAs would be appropriate at some point. I haven’t looked into funding yet but think that there would be some. Happy to coordinate in the US so efforts aren’t duplicated and are even more standardized.
@channel any thoughts on the ISBA abstract above?
And next GSC meeting is March 28th 😕 sorry this is taking so long...
*Thread Reply:* Yes, I added my comments in - can you see them?
*Thread Reply:* Yup thank you, already merged 🙂
Last thoughts on abstract? Otherwise I shall submit it tomorrow!
*Thread Reply:* only one minor grammar thing 🙂
*Thread Reply:* Nooooooooooooooooooooooo
*Thread Reply:* 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧 🇬🇧
*Thread Reply:* But thank you for looking through it 😄
Submitted yday! Will let you know on updtes!
@channel for everyone who has partcipated in drafting the current MInAS checklist, please can you check I've got your affiliations correct here!
https://github.com/MIxS-MInAS/MIxS-MInAS.github.io/pull/4
*Thread Reply:* There is a typo in my affiliation — great if you could update to: Peter D. Heintzman (Stockholm University :flag_se:)
Thanks, James!
*Thread Reply:* Dune!
Hi everyone!
Antonio and I finally had the initial meeting with the GSC.
To summarise, it sounds like we have their support but we are a bit weird in the way our data needs to be integrated, so we are going to have another meeting with the core GSC people to work out exactly in what form MInAS will go forward as (a whole checklist vs a package etc.).
In the meantime in whatever we will be creating, it seems we might end up being a 'guinea pig ' for them by being ones of the first to do a 'package first submission' where we basically submit our proposals in their new 'LinkML' format they have adopted (rather than they convert our table for them). So we need to be prepared for that.
They have also asked in preparation for the next meeting to have examples of datasets where aDNA would not fit in the existing checklists. So if anyone has any examples off the top of their head it would be great if they could post it here (e.g., can't provide the existing mandatory field of the local environment of the sample, as it's a museum sample that was deposited 100 years ago so the context has been lost).
Also in the future, all discussion and communication in the actual terms we propose will happen in the GSC GitHub page.
If anyone would like to join the next meeting with the GSC please let me know. But note it may be quite technical/philosophical
*Thread Reply:* If my schedule permits, I’ll be happy to join the next meeting.
Being weird and guinea pigs is like the aDNA credo at this point. Thanks for the update!
Hello again @channel!
Antonio and I met again with the GCS in a very illuminating meeting and has given us a much much clearer roadmap!
Some of the answer we got also helped us a lot with some of the common question that came up during our original meetings (wtf does conditional actually mean?!), so I'll summarise here:
Next steps is I will update the website to explain the new structure of the MInAS project, and transfer our proposed 'new' terms to the GSC github issue. I will maybe set up another meeting with all of us here to work out how we can start getting community feedback and funding for in-person workshops etc.
Hi @channel! With the changes above I think it's worth scheduling then next MInAS meeting 🙂
https://www.when2meet.com/?19936984-Ectmk (I've tried to increase the time, to see if we can get @Nasreen Broomand involved in this time, as her time zone has been awkward this time, based on the goodwill of Aida and childcare, but we will always go with majority vote)
@channel reminder of above 👆
@channel of the people who responded to the poll: we will meet at 15:30 CEST on Monday 22nd May!
Agenda:
• Review my website update for new MInAS proejct structure (if I have it done in time) • Assign people issues for adding ancient environmental package entires to the MIxS github repo • Collaboratively write some common slide material for presenting MInAS to our collective labs/groups for feedback (to use them as guinea pigs on how we can roll this out to the wider aDNA community)
Please note you can still join the meeting if you didn't respond to the poll - the more people the better!
We will meet primarily on gather.town, unless we end up with more than 10 people in which case we will move to Zoom.
If you want a google calendar invite please DM me your emails 🙂
@channel reminder we have a MInAS Meeting today at 15:30 CET! (in 2.5h!)
We will try meeting on gather here: https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community
But if we exceed 10 people we will switch to Zoom room ID:

Hello all, I am really sorry, something has come up and I won't be able to join 😞 Let me know if there is anything I can do to help via emails!
If I'm late that will be why
If you can't find me, open the Participant list, click on my name and 'follow me'
Website: https://www.mixs-minas.org/
My internet connection is really bad, I think I won’t be able to re-join the meeting
Don't worry we will summarise after :)(
@channel for those who couldn't make it today, meeting notes are here: https://docs.google.com/document/d/1fleZbCXOqXsybK7Hpq4ETv-v1HnN4C7CFjxDpvYd13Q/edit?usp=sharing for those who excused themselves but wanted tasks (or had joined via their phone), please see slide 11 of the slides here for their GitHub issue assignments (more isntructions will come soon): https://docs.google.com/presentation/d/1hyPVZlXJBhADbvh8-1se2JRFdVBz8mkRgT2Mm8tTb2U/edit?usp=sharing
For those in the meeting today (and @Ophélie Lebrasseur), the GitHub instructions for adding our assigned term proposals!
https://hackmd.io/@jfy133/HkValEYH3
Please let me know if anything is unclear
The complexted example: https://github.com/GenomicsStandardsConsortium/mixs/issues/546 (ignore the additional context)
OK I've added all mine, and so has @Mohamed Sarhan 🙂
*Thread Reply:* I’ll do mine soon, sorry! Crazy week
*Thread Reply:* No worries!
*Thread Reply:* working on this now, but I'm not able to add the env_package label. The little cog icon doesn't show up by the "labels" heading for me, sorry! I'll add my other three now and go back to try and figure out the label issue later
*Thread Reply:* also apologies if I missed a long discussion about this in a missed meeting but why is the "lib_concentration" in copies/µL as inferred by qPCR? Is this something people commonly do? I've never measured library concentration this way, we always report in ng/µL (and note whether it was measured by nanodrop or qubit, since that makes a huge difference). Also I'm assuming this is for already indexed libraries but it doesn't specifically note that, so I feel like clarifying that might be helpful?
*Thread Reply:* 1) ah yeah only I have permissions to do that at the moment, so you can skip that 2) I'm not a wet lab person, so we will need someone else to verify, but when I did my labwork during my masters, we only used nanodrop etc for modern DNA. It often wasn't sensitive enough for low biomass ancient DNA libraries, for which we used a tape station, which is more accurate as you measure the actual DNA molecules. I don't know if this is standard or not though... Also nanodrops are sensitive to non-DNA contamination, so that could make it difficult too. For us at least it's not indexed libraries but rather adapter ligated libraries (before amplification) because that should reflect roughly the amount of original DNA in the sample.
However this is a good test case for a discussion on the GitHub issue 😈! So copy over the original description, then in a new comment underneath the opening proposal say exactly what you've written above, and I can copy my reply too!
*Thread Reply:* Ok! Ill comment on the GitHub. I’d love to hear other people’s perspectives—I’ve been doing wet lab work for a long time but I’ve only worked in a few different labs so my experience doesn’t speak for everyone! Tbh yes nanodrop can be inaccurate but it’s consistent enough that a lot of labs do use it (and it’s nice to have something easy for a change where you don’t have to order any reagents lol). For me, when calculating concentrations for important things, I actually tend to trust qubit values over tapestation values. But that could just be me. In any case, different people use different methods (even methods that haven’t been mentioned yet) which is why I thought it might be good to include both concentration and method of measurement. :)
*Thread Reply:* Mine are up! And I added my thoughts on copies/ng per ul on Nasreen’s issue
*Thread Reply:* @Nasreen Broomand that's a good point, wanna poll the community on #general? How do you quanity your libraries etc.?
*Thread Reply:* I've added mines too!
*Thread Reply:* @Miriam Bravo you've missing a f ewthings (e.g. it's for MinAS)
*Thread Reply:* Also your backticks didn't see to work properly 🤔
*Thread Reply:* OK just @Meriam Guellil and @Ophélie Lebrasseur to go then we have all on board 🙂 (@Ophélie Lebrasseur just shout if you want me to very brielfly summarise the task over a call)
*Thread Reply:* Sorry I got hit by allergies really bad last week, will do it tomorrow, I haven't forgotten
*Thread Reply:* No worries!
*Thread Reply:* Hope you're feeling better now
*Thread Reply:* ok I added them, hopefully the right way. But I could not add the second label
*Thread Reply:* Awesome thank you very much!
*Thread Reply:* Only I of our group can do that at the moment
*Thread Reply:* @James Fellows Yates the issue about the negative will remain open, right? Do I need to do anything else about the issues I opened?
*Thread Reply:* Yes! There is discussion to be had in all of them
*Thread Reply:* FYI seems like we let a duplicate slip through
*Thread Reply:* Not necessarily
*Thread Reply:* We need to discuss a bit more precisely I think
*Thread Reply:* There is DNA storage and there is sample storage
*Thread Reply:* were making the point, that the variable could have an expanded definition
*Thread Reply:* but I agree that there is a difference
*Thread Reply:* Yes agreed. Maybe we can merge the two
*Thread Reply:* Maybe that would make more sense indeed 👍:skintone2:
*Thread Reply:* @James Fellows Yates Mines are now up too ^^ (hopefully I've done them correctly)
*Thread Reply:* Will check tomorrow!
*Thread Reply:* Looks good, thanks @Ophélie Lebrasseur!
Iv'e also made a sepaerate PR to make it easier, but waitnig on GCS now
Link: https://genomicsstandardsconsortium.github.io/GSC23-Bangkok/
@channel apparnetly our institute has an inhouse multimedia department that can also make logos!
But I need descriptive keywords of the project for them: I need ideas!
What do people think about switching phase two and three: https://www.mixs-minas.org/phases/
Us as the SpAAm community comes up with proposed submission packages (checklist plus extensions), send those out for review and then we can do term updates based on the feedback we get?
Any thoughts?
My logic being we can probably infer the package combinations relatively easily, then it's something more concrete we can send to people to review.
If we do you we can send them a sort of survey thing, and ask for comments, and if they provide them, and importantly pledge that they will use the package for their metadata tables was well as ENA/SRA uploads, they can go on the minas paper
How does that sound?
Also something else I want to do in our next meeting: I want to hear your motivations for contributing to the project
(to help me prepare the iSBA presentation, and hopefully appeal to more people why they should use it)
And we should set up a mailing list... Please can someone pester me to do this.
I also wonder if we could somehow track uptake... if we make a draft set of submission packages, then publish the kick a paper, then ask anyone who gives feedback/pledges to use the submission packages to also cite teh kick off paper (with a note saying this is the draft)... or do you think that would come off as a citation fishing...?
I would wait for the paper, first advertise at isba the ongoing project to get more people interested. Then expand the potential packages with input from new people and then get a draft of minas. After all this is in place we can start with the paper. If we go with a paper with a small part of the community we have the chances of not being well received.
Even if it's just a 'project announcemnet' thing rather than the actual paper?
This can done as a blog post in spaam
But no-one reads the SPAAM blog outside of SPAAM/ancient metagenomics is my fear 😕
OK but then maybe there is no point at all (the tracking thing would be useful but otherwise maybe it's not worth the time)
Let's prepare nice slides for your talk and draft a blog post. We can publish at the anvio blog as well and mirror it at spam. Then spread it through some of my friends with large twitter audience
@channel we have most of the ancient package terms on github now (just couple from Ophélie when she's available again left)
So we should arrange the next meeting.
Agenda will be:
- Making submission package proposals (new phase 2)
- Finalizing the lab-based 'advertising and feedback' slides Please fill in form here: https://www.when2meet.com/?20269802-kZcu2
Also @Antonio Fernandez-Guerra is investigating how to set up a mailing list to help get more people onboard out of SPAAM (E.g. slack-allergic PIs 😉 ). We will let you know once we have a solution
@channel reminder to fill in the when2meet!!!! ☝️ only have 3 responses
@channel (sorry for the pestering), and in particular @Antonio Fernandez-Guerra (!!!) last chance ☝️ I will shut tomorrow afternoon
*Thread Reply:* I only have Tuesday available, but doesn’t seem the best day for the others.
*Thread Reply:* Just vote 😉
@channel next meeting: Friday 23rd 15:00_16:00 CEST (Berlin time)! As always send me google account if you want a google event invite
*Thread Reply:* Could you send me an invite blevinske1@gmail.com
Agenda:
• Check all github issues added/updated • Define draft submission package proposals • Set lab presentations targets, and finalise slides/procedure
*Thread Reply:* Regarding replies to Github issues, should we handle them before the meeting or discuss the optimal reply/plan during the meeting together first so we are all on the same page?
*Thread Reply:* The purpose of the issues now is the place to track discussions (so we have a history), so ideally leave comments there 🙂
(@Ophélie Lebrasseur let me know if you need help with teh github issues)
*Thread Reply:* Thank you @James Fellows Yates, I'll get on it wednesday and let you know how it goes 🙂
@channel last reminder this meeting on Friday, remember anyone can join even if you didn't respond to the poll!
Hi @channel We have our space at #minas in the GSC Slack. Please use this invitation link (https://join.slack.com/t/gsc-vpc4453/shared_invite/zt-1xkho921f-NeTTzLUS~cSF4HBkOSXFQg) to join the channel. The mailing list will be available soon. We are updating the website to reflect these changes. Please share this information and link to anyone interested, from now on, we will be using the GSC related communication channels for our project. Thanks!
Lets try gather, and if we fill up we switch to Zoom @channel
*Thread Reply:* Sorry something came up last minute so I will have to skip today
*Thread Reply:* Ok no worries
So gather: https://app.gather.town/app/PlXjb0deog0B4JCq/spaam-community Meeting Agenda/notes: https://docs.google.com/document/d/1dmYUin_3Fz41mPplpZScMimr7yR0uiy1d_PrQmxQbGk/edit?usp=sharing
@Nasreen Broomand will you be able to join?
@channel what would people think about setting up a monthly meeting time (e.g. every 4th Tuesday of every month) rather than polling each time?
That works for me 🙂 In general, I can't do every last Tuesday of every month but apart from that, I should be good
(doesn't ahve to be tuesday, just an example)
If everyone agrees, please let me know what day you generally have more time on:
1️⃣ Mon 2️⃣ Tue 3️⃣ Wed 4️⃣ Thu 5️⃣ Fri
*Thread Reply:* Just a reminder that I can only make it to certain times because I’m on pacific time. Thanks! :)
*Thread Reply:* :salute: absoultely it's in mind!
@channel First Thursday of every month at 15:00_16:00 CES(S)T (Berlin Time)?
Yes: 👍 No: 👎
*Thread Reply:* I can not do at 15:00
*Thread Reply:* ....sooooooo what time can you do? 😬 Note we need to account for @Nasreen Broomand
*Thread Reply:* or 14:00 or then late evening, I pick up my kid at 15:00
*Thread Reply:* @Nasreen Broomand 14:00 is too early for you right?
*Thread Reply:* but go for 15:00 and i might be able to join depending on Marina’s lab work.
*Thread Reply:* 14:00 is 5 am for me 😬 I’d rather avoid 5 am meetings but can make it if necessary
*Thread Reply:* Or we go 8AM CEST which is 23:00 for you I think? 😆 are you a night owl or morning person 😬
*Thread Reply:* I could do either! 5 am is probably better because I’ll have time afterwards to do tasks (whereas if I have a meeting at 11 pm I will just put it on my to-do list) but they’re pretty much the same to me! So whatever works for the group as a whole
*Thread Reply:* I promise not to ask you to turn on your webcam ❤️
@channel (compromise between @Nasreen Broomand and @Antonio Fernandez-Guerra!) First Thursday of every month at *14:00_15:00 CES(S)T* (Berlin Time)?
Yes: 👍 No: 👎
@channel sorry this is last minute!
I am presenting today to the pathogen groups here in Leipzig. Unfortuantely I've been a bit overwhelmed workwise (damn summer school :shakes_fist: ), and was only able to finish drafting the template today:
https://docs.google.com/presentation/d/1OvZkKV5cmX6CxfBU8fqaePAhT9fueij_Rbzalfqpndg/edit?usp=sharing
Please give feedback if you can! But we can still update teh template after 🙂
One major change is on how we can try and gather feedback. I think going through 100 terms would be much to dry and boring and will put people off, so I had the idea instead of trying to make it a more pratical exercise.
- You present the project and then given them the relevent (draft) submission checklist
- The people have 2-3 weeks to try filling in the checklist with maybe 5-10 samples from their last paper or project
- Then they bring this back with them after the 2-3 weeks ,and then you have a round table discussion on the pain points etc What do you think about that approach? In the future maybe we can bribe people with stickers etc as well (not just a possible nature biotech project or something)
Hello James, I've had a quick look (can dedicate more time towards the end of the week), but at first glance, I think it works very well. And I like the idea of asking people to have a go with their own sample, as every person will most likely have different type of samples, especially across labs and geographical regions, so I think it'll give us a good overview of the applicability of MInAS. My only comment regards the first two discussion points. I guess these are more general questions thrown to the floor to get an overall feel rather than actual discussion points?
*Thread Reply:* Yes, it's more of a hands up polling thing
*Thread Reply:* I'll change it to question target than discussion
*Thread Reply:* Done, do you think that is better to Indeed get the feeling of the floor line you said?
*Thread Reply:* I think so personally. It gets people into the themes through two main questions, and I feel the second question will generate straightforward answers that will directly feed into the whole purpose of MInAS (my guess is it might even lead to a domino effect as there are a lot of categories that are not present for archaeological samples and once you start listing these categories, they just come tumbling down ^^). And as these questions are more general, it means not too much time is spent prior to explaining MInAS and also reduces repetitions. These are only my thoughts 🙂
*Thread Reply:* That's a good point about straightforward answers... I've changed the 3 into 4 questions, and they are all Y/N polls now (so hand raising) rather than needing to discuss stuff... do you think that would be better so we don't repeat so much?
*Thread Reply:* I like it! Slide 6 I think has a couple of typos. And I think for slide 5 we can still allow for some general observations. I guess that depends on how active the participants are ^^
*Thread Reply:* I fixed the typos!
I think it's a very nice presentation! I just have a comment about the order of the slides; switch the slide 14 (Ultimate goal..) before the Slide 18, and the the Slide 16 (Long term goal..) after the 19 (Second goal). It's just that I was little bit confused to see the goals in that order..but I don't know..what do you think?
*Thread Reply:* Uhh that's a good point, phasing of ultimate and long term probably doesn't make sense indeed! I guess better would be immediate and long term
I can start translating it into spanish!
*Thread Reply:* Let me know if I can help @Miriam Bravo
*Thread Reply:* sure, podemos dividirnos las diapositivas, si quieres selecciona con las que quieras comenzar 🙂
*Thread Reply:* si quieres yo hago las primeras 16 diapositivas y vos las siguientes 16, te parece?
*Thread Reply:* Acá están las primeras 16 diapositivas @Miriam Bravo https://docs.google.com/presentation/d/1-_s-osfo9kE90pFlHsn83KHTixxcxBCnZv9m350PYKA/edit?usp=sharing
So update from yesterday:
• Meeting went well, the template slides need a little more finesse (a bit of duplication, which I've already partly addressed) • The polling was a good ice breaker, and people I think appreciated having a specific task they can do in their own time first - before getting into discussions • We will see how it has worked in 3 weeks But I encourage others to already start planning/using the slides in their own labs so we can see how well it works too. Happy to chat if you have any more questions about how I went about presenting it!
I will also set up the monthly meeting in my google calendar - if you want a google invite please let me know - but otherwise: First Thursday of every month at 14:00_15:00 CES(S)T (Berlin Time)?
That would technically be today, but I imagine thsi is too short notice 😬
Woop woop, well done. that's great feedback 🙌😊 I'll try and see if I can organise a presentation here in CAGT, and one online with the PalaeoBARN and Laurent Frantz's group in Munich. Do you need me to translate the slides in French? As for the meeting, I've added the dates to my calendar, and am free for a meeting today
DEfinitely if the french would help!
@channel a reminder to please join the GSC workspace! https://www.mixs-minas.org/get-involved/!
I'm posting some possible logo options there we need to vote on 🙂
Also I've realised (or rather Kelly made me realise) that this first 'regular' MInAS meeting I won't be able to do as it will be the SPAAM Summer School
How would everoyne think about moving it back one week (10th August) @channel? Or is most people gonna be away/holiday?
That is the only week I am on holiday, so I won't be able to make it
I realised I have a medical appointment for the original date (3rd August) that I booked several months ago and really can't change, and then I am away the week after (10th of August), so will be able to join from September onwards :/
OK no worries, last call @channel about a meeting on the 10th? If not we can just postpone meeting until SPAAM5/ISBA 🙂
I would postpone it until spetember
@channel small update over on the GSC slack 😄
Hi @channel! Just to let you know @Kelly Blevins has agreed to join the main coordinator team with Antonio and I, to initially help tide things over while I'm on leave and then help coordinate the project next year. If you have any questions (e.g. you want to start getting feedback from your own groups, or help with translations of the feedback slides), you can also speak to her!
Hi, all! Our next minas meeting dates and times are below for the rest of the year. At the October meeting we will go over the feedback we’ve gotten, including from ISBA and at lab group meetings. We will also discuss how you can run your own minas meeting in a lab group near (or far from) you. Hope to see some of you in Tartu!
5 October 2PM CEST 2 November 2PM CET 7 December 2PM CET
@channel if you're at ISBA10 I have MInAS stickers! Come find me and I'll give them to you 😄
*Thread Reply:* @Kelly Blevins we should also meet for a bit to chat and I can give you half of them
*Thread Reply:* @James Fellows Yates Can you pass me one lol
*Thread Reply:* Or ask me to suspiciously pass it along the row in the middle of a talk
*Thread Reply:* Sure! I’ll find you at the coffee break
Hi, @channel! We were meant to meet next week 5 October at 2PM CEST, but the following weeks works best for some people. Please fill out the poll to help us decide if we should move October’s meeting.
Hi guys, I'm sorry, I'm at ICAZ AGPM that week and away next week. I can make the week of the 16th though
Apologies for the date switch, everyone. We are going to move the next minas meeting to Thursday 12 October at 2PM CEST. After that, the meetings will be the first Thursday of the month.
Hi, @channel! Reminder that we have a meeting today at 2PM CEST:
MInAS recurrent meeting (first Thursday of the month): https://ucph-ku.zoom.us/j/61373395453?pwd=V0ZyYVVSa2x6Tm0wVUFsY3RwdjFpdz09
Agenda: https://docs.google.com/document/d/1HgpSolHyMtc0lAWnFIArH3OGCX4d0qyq17ZsBg-z23g/edit?usp=sharing
Hi, @channel. Apologies but we are rescheduling our next MInAS meeting for NEXT Thursday 9 November at 2PM CET. There will be no meeting this week due to scheduling conflicts for all coordinators. Sorry for the inconvenience, and thanks for your understanding.
Hello @Kelly Blevins, thank you for letting us know! I will be away next week and sadly won't be able to join, but as always, let me know if I there is anything I can do to help.
Hi, @channel! See you in 1.5 hours for the next MInAS meeting!
Zoom: https://ucph-ku.zoom.us/j/61373395453?pwd=V0ZyYVVSa2x6Tm0wVUFsY3RwdjFpdz09 Agenda: https://docs.google.com/document/d/1LBZy6DW7W9aFXlR4NbfIN2NHQfrByEK7L-Kpn9-fAvU/edit
Hi all, I’m so sorry I missed it! I had three vaccines yesterday so I’m feeling kinda crappy and just completely forgot.
hello! Sorry for missed that too, I didn't have electricity in my place 😞
Hi, @channel Apologies for the late notice, but because of chaotic end-of-year workloads, we’ve decided to postpone this week’s MInAS meeting until next year. Happy holidays, everyone! 🎄🍾
@channel meeting starting 6 minutes ago! See the GSC slack channel 😄









 }
}