Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
New preprint: https://www.biorxiv.org/content/10.1101/2020.04.20.050369v1.article-info

Does anyone know the authors and could ask for the supplement? It seems a lot of the information is there...
*Thread Reply:* WAs it originally tweeted by someone?
*Thread Reply:* Yep, first author tweeted it. His handle is at the bottom of the bioRxiv page for the paper
Could we do some shameless self-promotion with our latest paper? https://www.frontiersin.org/articles/10.3389/fevo.2020.00084/full
@Sterling Wright has joined the channel
https://www.nature.com/articles/s41467-020-16502-3
*Thread Reply:* Very honest too, props to htem!:
From the bulk bone samples, we did not find any evidence of cross contamination between samples,
however, in one sample (HCB23_B) we identified significant contamination from fauna of New Zealand,
which could stem from samples processed in the same laboratory for a different project. To identify the
source of this contamination, we re-extracted and amplified the bone powder from HCB23_A and HCB23_B,
which confirmed the presence of significant DNA contamination in the bone powder of both of these
samples. Next, we returned to the original bulk bone samples, sampled and processed another 2x50 bones
each. These samples contained only species from Texas and were comparable to other samples from the
surrounding layers. Hence, we conclude that the bulk bone powder from the two subsamples of HCB23, was
contaminated during the bone grinding stage. Most likely, this contamination stems from the reuse of a
grinding pot that had not been cleaned properly. To confirm that this contamination event was a single
incidence, we processed two ‘grinding blanks’ in which 15 mL of ultrapure water was run in the ball mill under
the same configuration as the bulk bone samples. After grinding, the water sample was concentrated to 500
µL in an Amicon®Ultra-4 Centrifugal Filter (Millipore) and processed as a bulk bone sample. From these
samples, we only identified background contamination from Homo sapiens
(I wonder how their data could be applied to PIA @Becky Cribdon? Looks like they take a similar-ish approach as they use BLAST)
Thanks for highlighting this, James. If they used BLAST, they should be able to use PIA. Looks interesting...
Hmm, they sequenced using metabarcoding. It would certainly be possible to analyse their data with PIA, but it's been developed for shotgun sequencing, so I'm not sure what effect it would have on the output. I've asked a friend who has done this sort of thing; it's a good question.
*Thread Reply:* Yeah, I realised that after too. Too used to sedaDNA being shotgun
*Thread Reply:* I'd like to see some more of it myself! Friend says PIA would probably do the same sort of thing on metabarcoding data, but it's not ideal.
*Thread Reply:* Did they say why it's not ideal? Not enough sequence diversity or somethin?
https://www.frontiersin.org/articles/10.3389/fevo.2020.00218/abstract
@Anneke ter Schure has joined the channel
Not directly related to metagenomics methods per se, but for those working on oral microbiomes: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-020-06810-9 (via @ivelsko)

While we don’t have a session directly addressing data reporting, this is something worth working towards with ancient metagenomics https://www.biorxiv.org/content/10.1101/2020.06.24.167353v1.abstract?%3Fcollection=

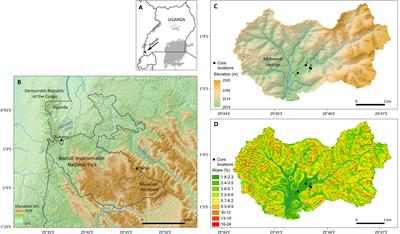
Some sedaDNA in a really fascinating site it seems!
☝️ maybe a good basis for SPAAM
STROBE-metagenomics: a STROBE extension statement to guide the reporting of metagenomics studies Summary The term metagenomics refers to the use of sequencing methods to simultaneously identify genomic material from all organisms present in a sample, with the advantage of greater taxonomic resolution than culture or other methods. Applications include pathogen detection and discovery, species characterisation, antimicrobial resistance detection, virulence profiling, and study of the microbiome and microecological factors affecting health. However, metagenomics involves complex and multistep processes and there are important technical and methodological challenges that require careful consideration to support valid inference. We co-ordinated a multidisciplinary, international expert group to establish reporting guidelines that address specimen processing, nucleic acid extraction, sequencing platforms, bioinformatics considerations, quality assurance, limits of detection, power and sample size, confirmatory testing, causality criteria, cost, and ethical issues. The guidance recognises that metagenomics research requires pragmatism and caution in interpretation, and that this field is rapidly evolving.
@Antonio Fernandez-Guerra has joined the channel
New pathogen genomes: https://bmcbiol.biomedcentral.com/track/pdf/10.1186/s12915-020-00839-8
“The archives are half-empty: an assessment of the availability of microbial community sequencing data” https://www.nature.com/articles/s42003-020-01204-9
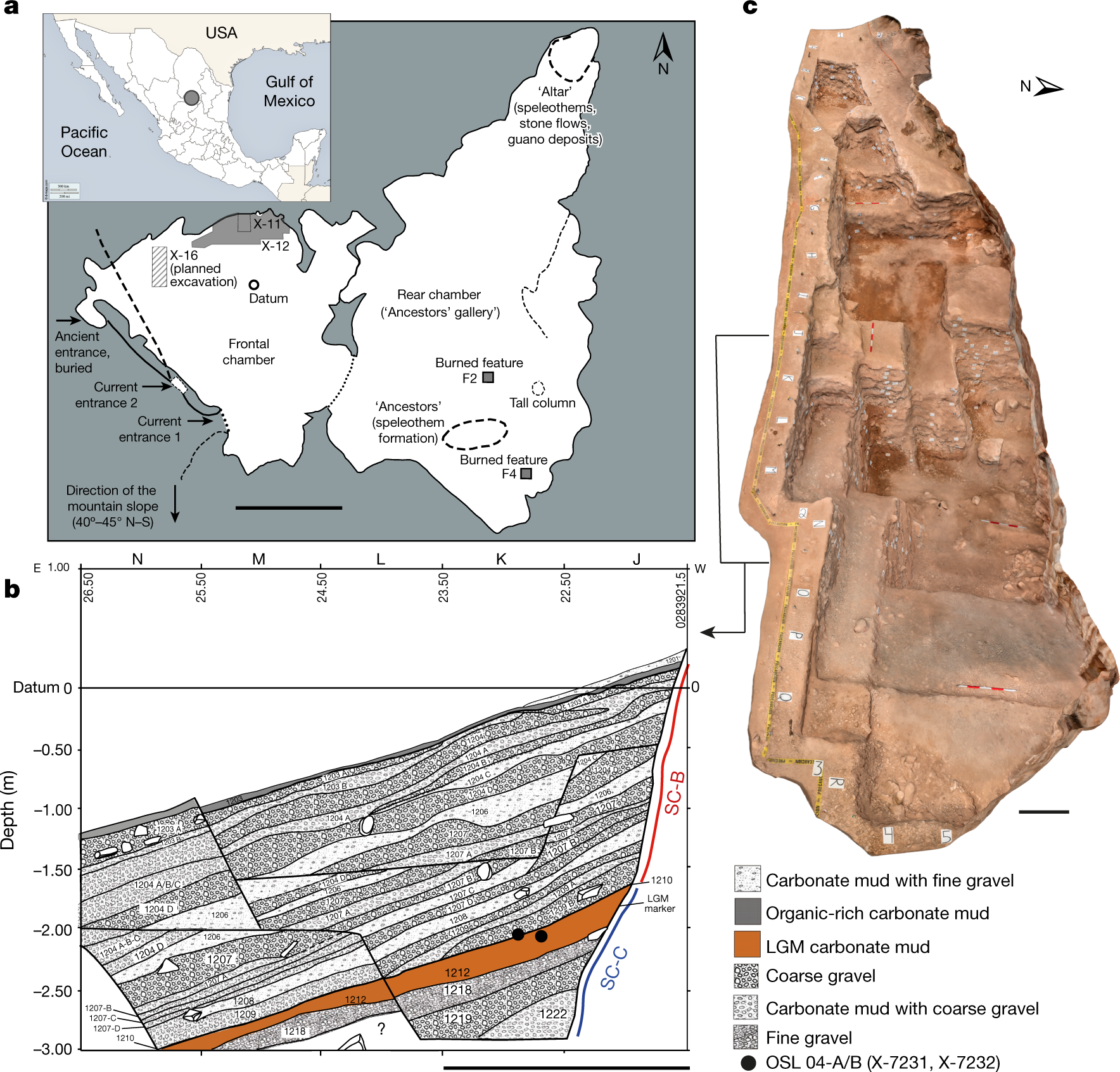
“Optimizing extraction and targeted capture of ancient environmental DNA for reconstructing past environments using the PalaeoChip Arctic-1.0 bait-set” https://www.cambridge.org/core/journals/quaternary-research/article/optimizing-extraction-and-targeted-capture-of-ancient-environmental-dna-for-reconstructing-past-environments-using-the-palaeochip-arctic10-baitset/BDC98B87B7A68968FA99204512D8ECFE
*Thread Reply:* Could not see any raw data released, unfortunately - but a great paper for those working with ancient environmental DNA!
Saidi-Mehrabad, A., Neuberger, P., Cavaco, M. et al. Optimization of subsampling, decontamination, and DNA extraction of difficult peat and silt permafrost samples. Sci Rep 10, 14295 (2020). https://doi.org/10.1038/s41598-020-71234-0
I guess sedaDNA is all the range atm ;
https://www.annualreviews.org/doi/abs/10.1146/annurev-micro-012520-072314?af=R
https://pubmed.ncbi.nlm.nih.gov/32943086/

*Thread Reply:* Poop-gen only though 😢
Ancient viruses from Japanese teeth? https://www.nature.com/articles/s10038-020-00841-6
Multi-omic detection of Mycobacterium leprae in archaeological human dental calculus: https://royalsocietypublishing.org/doi/pdf/10.1098/rstb.2019.0584
- and a whole issue 😱: https://royalsocietypublishing.org/toc/rstb/2020/375/1812
Cool paper, but no raw data released unfortuntely: "Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau" https://science.sciencemag.org/content/370/6516/584
*Thread Reply:* Or still embargoed?
*Thread Reply:* Only human mito consensus sequences released. No mention of raw data:
"Data and materials availability: ... The new mitochondrial consensus files reported in this paper have been deposited in the Genome Warehouse in National Genomics Data Center (24), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number PRJCA002765, which is publicly accessible at https://bigd.big.ac.cn/gwh...."
*Thread Reply:* Poop-gen 😏 .
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7585846/

De novi assembly and binning of C20 bacterial genomes from dental calculus
https://www.nature.com/articles/s41559-020-01351-6

https://www.biorxiv.org/content/10.1101/2020.11.14.382994v4

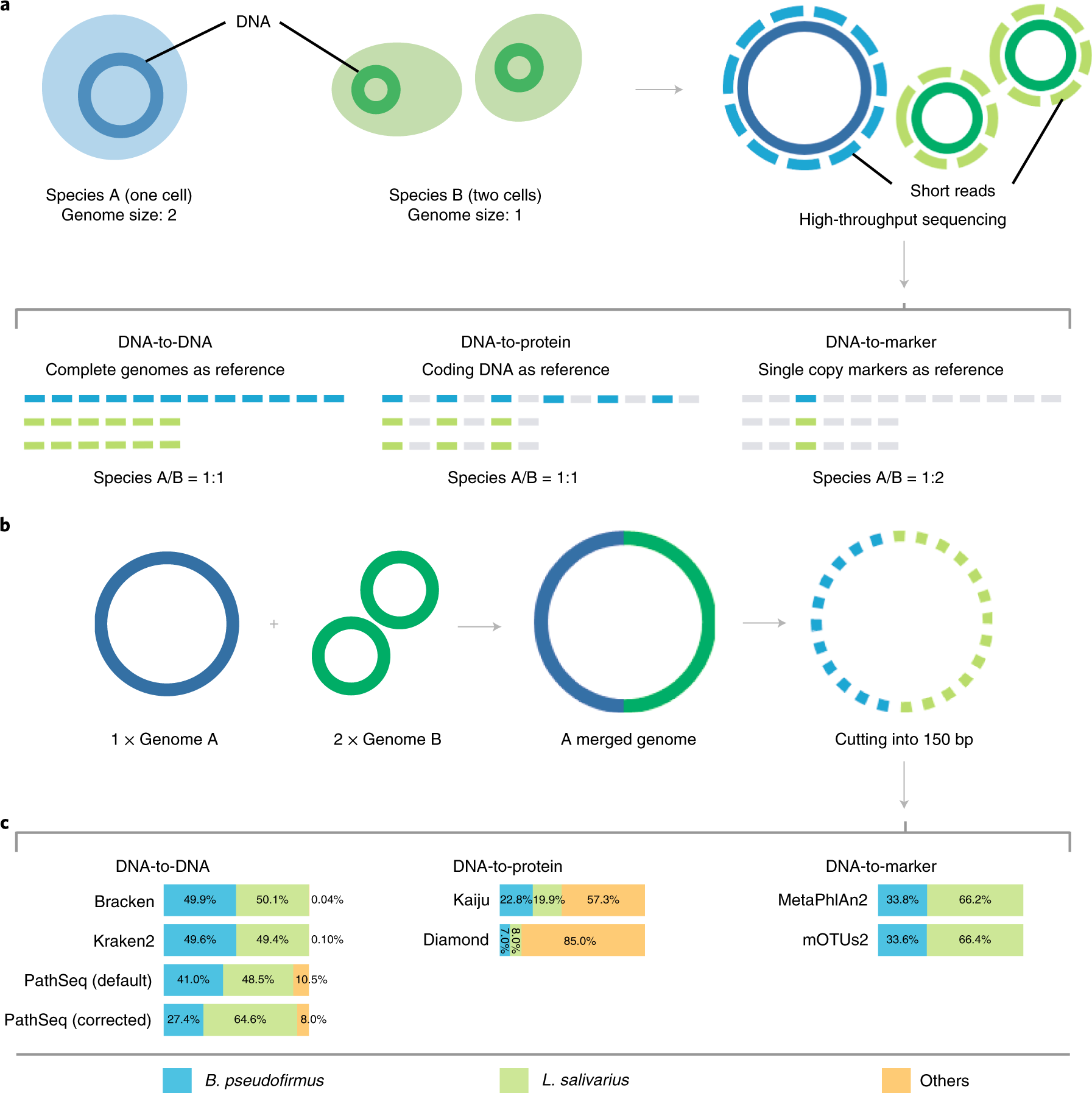
". We find that microbiome data analysis based on sequence abundance will underestimate (or overestimate) the relative abundances of microbes with smaller (or larger) genome sizes. This will fundamentally affect differential abundance analyses and other analytical methods that rely on accurate counts in their input contingency matrix"
"Taken together, these analyses emphasize the importance of differentiating between relative sequence abundance and relative taxonomic abundance in metagenomic profiling. Ignoring this distinction can potentially underestimate the relative abundance of organisms with small genome sizes."
Some more formalised pangenome graphs for on-humans!
https://www.biorxiv.org/content/10.1101/2020.11.12.380378v2

Why are the authors silent on this paper? It's a great read! https://www.sciencedirect.com/science/article/pii/S1040618220307746

*Thread Reply:* Because they didn't tell us as first they uploaded an uncorrected version... and it's meant to be OA but they won't reply to us about how we can pay for it 🤦 .
*Thread Reply:* But I see the final version is live, so will tweet it in a bit 😅
*Thread Reply:* Congratulations! Just reading the intro now (and checked out the figures) and I love it!
https://www.biorxiv.org/content/10.1101/2020.09.28.316869v1

Series of responses to ancient small pox (issues of applying modern assemblies to ancient DNA) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7716422/

If anyone is curious how Archaeogenetics at MPI-SHH samples teeth for pathogen DNA, we just made our sampling protocol in step-by-step format open access and citable on protocols.io! https://dx.doi.org/10.17504/protocols.io.bqebmtan (Co written with SPAAM members @Gunnar Neumann and @aidanva!)

@Gunnar Neumann has joined the channel
Some group and self-promotion. All and any comments welcome! https://www.biorxiv.org/content/10.1101/2020.12.22.423960v1

*Thread Reply:* Woohoo! Finally! Excited to read it!
Comparison of lab decontamination procedures of dental calculus
https://www.researchsquare.com/article/rs-136836/v1

Coauthored by @Sterling Wright !
*Thread Reply:* Small suggestion @Sterling Wright you may want to rename your GitHub repo (if not too late), 'decontam' is the same name as a (very good!) R package: https://github.com/benjjneb/decontam
Which has a statistical method for predicting likely lab contaminations.
*Thread Reply:* You might not want people to get confused, and I've seen some fights over bioinformatic tool names.... (Look up falcon ancient DNA metagenomics)
*Thread Reply:* Thank you for the heads up James! I’ll make sure to get with the co-authors and change it ASAP
Something similar what we would like to do for metagenomics but for c14 dating!
https://twitter.com/AmineNamouchi/status/1352230217776037888?s=19
Maybe something interesting to replace MultiVCFAnalyzer? Has anyone had any experience with this?
In case anyone isn't in Twitter, the first paper coming spinning out of the SPAAM community is now out! https://twitter.com/ScientificData/status/1354059031883747330?s=19
New dog poop paper: https://www.nature.com/articles/s41598-021-82362-6#Sec11
*Thread Reply:* @Ele could you add the paper as an issue to the 'Dir
Although I hope they've uploaded the raw data and didn't remove the <75bp as they do in the analysis :/
*Thread Reply:* Just added it as I had a spare moment and so we don't forget!
a primer on aDNA including the first citation for AncientMetagenomeDir!!! Written by PIs of a lot of aDNA labs from around the world! Free pdf link courtesy of @Christina Warinner!
Can you share a PDF? Can’t download from that link
@Christina Warinner has joined the channel
Hi All, does anyone have access to this and could share the pdf? “How reliably do metagenome-assembled genomes (MAGs) represent natural populations? Insights from comparing MAGs against isolate genomes derived from the same fecal sample” https://aem.asm.org/content/early/2021/01/12/AEM.02593-20
“Microbiome Metadata Standards: Report of the National Microbiome Data Collaborative’s Workshop and Follow-On Activities” https://msystems.asm.org/content/6/1/e01194-20
*Thread Reply:* Pet peeve: 'national intiative for xyz'... national for WHO!?
*Thread Reply:* It sucks for LIMS too 'Natural History Museum' x1000000
*Thread Reply:* (vi) Establish a certification of “compliance.” Despite the significant efforts already invested in defining minimum standards for microbiome data, such as the Minimum Information about any (x) Sequence (MIxS) packages (39), important work remains to ensure that the various standards and ontologies are interoperable and easily accessible to the research community. This entails working with researchers to identify metadata attributes that are valuable for data reuse within their respective communities, and defining community-specific benchmarks. Establishing a “certification of compliance” based on these benchmarks would enable designation of data sets ready for reuse, which encourages inclusion in follow-up studies and enhances their citation metrics (see section i above).
@Antonio Fernandez-Guerra ☝️
One for the calculus people: “Resilience of the oral microbiome” https://doi.org/10.1111/prd.12365 … “Diet, with the exception of excessive and/or frequent consumption of fermentable carbohydrate or supplementation with nitrate, has minimal impact on the composition of the oral bacterial community.“. This is quite a different perspective compared to stool studies
nf-core/eager paper is out!
Fellows Yates JA, Lamnidis TC, Borry M, Andrades Valtueña A, Fagernäs Z, Clayton S, Garcia MU, Neukamm J, Peltzer A. 2021. Reproducible, portable, and efficient ancient genome reconstruction with nf-core/eager. PeerJ 9:e10947 https://doi.org/10.7717/peerj.10947
Any suggestions for improving the metagenomic component welcome! Development doesn't stop here 🚀
*Thread Reply:* Ahh, I've been trying and failing to use EAGER - this could be the answer to all of my problems!! Very excited to try this, and congratulations on publication.
*Thread Reply:* Do you mean eager1?
*Thread Reply:* And thanks :)
PyDamage: automated ancient damage identification and estimation for contigs in ancient DNA de novo assembly - https://www.biorxiv.org/content/10.1101/2021.03.24.436838v1

Hi! I was wondering if someone has access to this paper: https://www.sciencedirect.com/science/article/abs/pii/S0966842X21000445 and wouldn't mind sharing the pdf??
*Thread Reply:* https://paperpile.com/shared/hV5rn8

May worth checking this method out: https://www.nature.com/articles/s41587-021-00861-3
https://twitter.com/NikolayOskolkov/status/1378021308311683072?s=19
Really nice blog post from @Nikolay Oskolkov on being careful when reporting detection of pathogens in aDNA data!
@Nikolay Oskolkov has joined the channel
this is a very good complement to @Nikolay Oskolkov post https://instrain.readthedocs.io/en/latest/important_concepts.html
https://science.sciencemag.org/content/early/2021/04/14/science.abf1667

https://www.nature.com/articles/s41586-021-03532-0
*Thread Reply:* @Katerina Guschanski this is the one I was talking about. I've not read it yet so can't say if it's the thing I was talking about
*Thread Reply:* Super! Thanks. Will have a look
Have you seen this one? It’s from a guy I know…. Very cool work! https://www.pnas.org/content/118/20/e2021655118
I dunno, I wouldn't trust him. He is pushing a veganist agenda that is a part of the 'great reset plan' apparently.

Now I read all the comments below! They are even better than the article! hahaha So good… You must be proud James….
@channel Does anyone have access to this paper and could share it with me? https://www.cell.com/trends/microbiology/fulltext/S0966-842X(20)30327-9?returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0966842X20303279%3Fshowall%3Dtrue|https://www.cell.com/trends/microbiology/fulltext/S0966-842X(20)30327-9?returnURL=[…]vier.com%2Fretrieve%2Fpii%2FS0966842X20303279%3Fshowall%3Dtrue
Might be relevant for others as well 😉
*Thread Reply:* Haha, @Abby Gancz was 2 seconds faster than me 🙂!
*Thread Reply:* Thank you!!! So speedy :donut_parrot:
This might be relevant to people who are trying to use e.g. decontam to remove lab contamination. (Cc @Markella Moraitou @Adrian Forsythe @Katerina Guschanski in particular, given the recent discussions)
For any American**/SRA users: this might be useful for generating standardised (following GCS) metadata tables for your studies: https://metagenote.niaid.nih.gov/
Attempt at Malaria capture: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8202054/

Ozga, A. T. and Ottoni, C. (2021) ‘Dental calculus as a proxy for animal microbiomes’, Quaternary international: the journal of the International Union for Quaternary Research. doi: 10.1016/j.quaint.2021.06.012. PDF: https://paperpile.com/shared/jt464e

https://www.nature.com/articles/s41586-021-03675-0

https://www.biorxiv.org/content/10.1101/2021.06.28.450199v1

Via retweet from @Zandra Fagernäs
Congrats to @Maxime Borry @Alex Hübner @Christina Warinner
https://peerj.com/articles/11845/
Big congrats on the pre-print @Zandra Fagernäs and co-authors! It is such a cool and important study! Will be our citation staple 🙂 https://www.biorxiv.org/content/10.1101/2021.08.16.456492v1

Congrats to @Jaelle Brealey (or is it @Jaelle Brealey?) And @Katerina Guschanski!
https://www.sciencedirect.com/science/article/pii/S096098222101112X

https://genome.cshlp.org/content/early/2021/09/03/gr.275777.121.abstract
A potentially cool alpha diversity metric to implement in microbiomes characterizations
https://www.nature.com/articles/s41586-021-03798-4
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009886
Ancient Chinese S. enterica
Congrats to @Lena G @Kun Huang!
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-021-01132-8

137 Hep. B virus genomes, with the co-author list including @Maria Spyrou @Felix Key @Gunnar Neumann @Kerttu Majander @Susanna Sabin @Megan Michel @Betsy Nelson @Marcel Keller @Karen Giffin and probably a bunch of other people I missed because going through 171 authors is a tad too much
https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-021-01120-2

*Thread Reply:* ANOTHER! Could you add as an issue to AncientMetageomeDir bitte? 🙏
*Thread Reply:* I did it
https://www.sciencedirect.com/science/article/pii/S0960982221012719

And another! from @Mohamed Sarhan and @Kun Huang!
@Mohamed Sarhan has joined the channel
From @Bjorn Bartholdy! https://www.biorxiv.org/content/10.1101/2021.10.27.466104v1.abstract?%3Fcollection=

https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001421
Careful of finding 'pathogens' in Metagenomic screening ;)
Ha-ha, this is brilliant @James Fellows Yates! Nowadays it is so easy to find pathogens by doing absolutely legal (or at least justifiable) manipulations with reference databases :))) so that report is not surprising ;)
Or basing the identification on a partial mapping and a metagenomic hit...
*Thread Reply:* I sense a sub-tweet here 😉
Exciting multidisciplinary work by @Meriam Guellil and co-authors
https://www.nature.com/articles/s41598-021-98214-2
We might have seen this in pre-print, but here's the final version https://www.nature.com/articles/s41591-021-01552-x
*Thread Reply:* Oooohhhh "presented as an editable table for inclusion in supplementary materials"
*Thread Reply:* Definitely need to read this
*Thread Reply:* Now to find time to actually read it
*Thread Reply:* At this stage in your career, you should be training to just absorb papers, no more time wasted "reading"
*Thread Reply:* Ok, find time to absorb 😆
*Thread Reply:* I don't think it matches what I immediately want for the standardised metadata sheet, but certainly it's something that we should aim for in the future 👍
*Thread Reply:* basically it tells you 'how to write your paper' for a specific type of microibome study, which might not necessarily work for our field at the moment (exactly). But certainly we could adopt some of these things.
It's just a shame they've saved it as shitty DOCX/XLSX files and not something under version control, so it would make it harder to fork and modify
*Thread Reply:* Yeah it really is a check list and not a template for what to upload
*Thread Reply:* The MIxS stuff is what we should aim for initially
*Thread Reply:* (and tools to make that easier)
https://www.mdpi.com/2079-7737/10/12/1324

From @Toni de Dios Martínez et al!
And also this preprint from @Katherine Eaton
https://www.researchsquare.com/article/rs-1146895/v1

https://www.sciencedirect.com/science/article/pii/S1040618221005735

https://www.nature.com/articles/s41467-021-27542-8
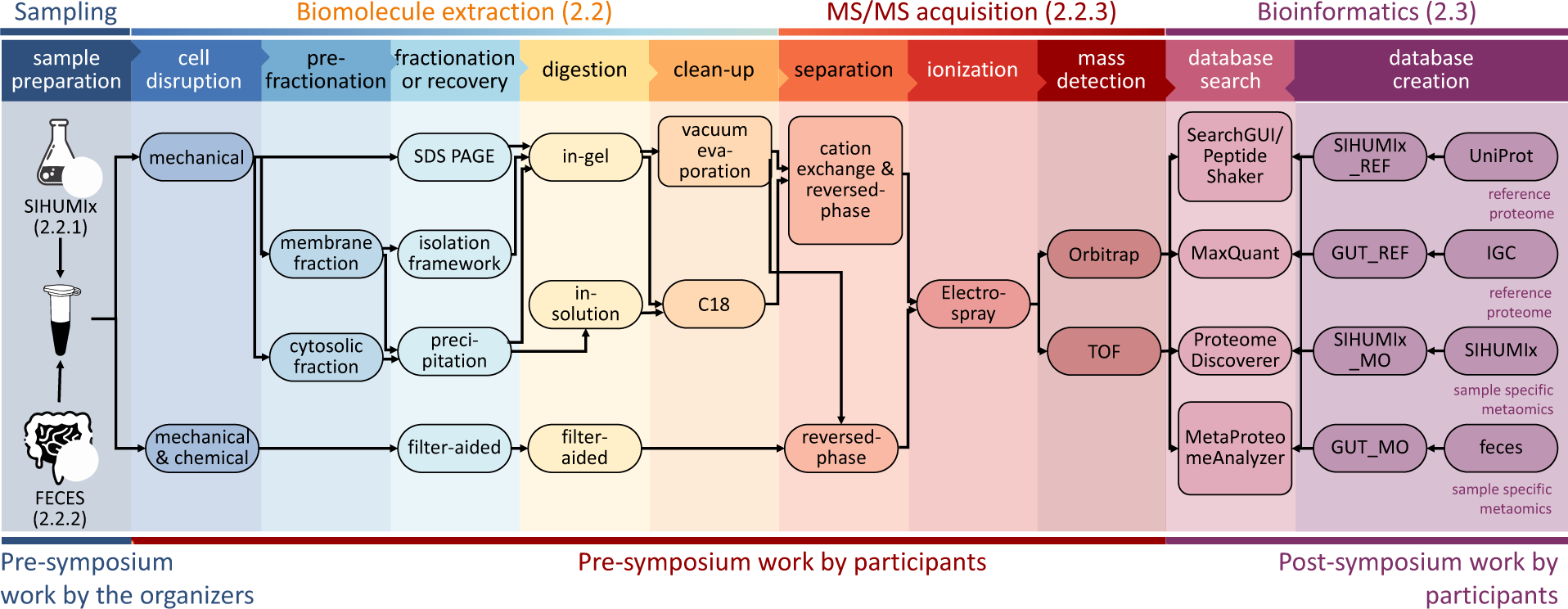
Maybe a good study to replciate for ancient meagneomics / ancient proteomics ☝️
https://journals.asm.org/eprint/X56YB6KCQVCCDS4Q9F3N/full
The legendary, mythical, fabled <#CPHECT30A|spaam2-open> opinion paper is now published in mSystems!
Thank you to everyone who attended :spaam: SPAAM2, and those who also gave further contributions, comments, and feedback for this paper. It was certainly more difficult than I expected since we proposed the idea - reflected in our challenges to pin down a single topic to talk about - however overall I think this reflects the exciting potential and movement happening in our field!
Let's hope the paper gets some interest from modern metagenomicists and open up some discussions!
I would like to also finish with a personal BIG shout-out and eternal gratitude to @Clio Der Sarkissian @Anna F. @Alex Hübner @Åshild (Ash) and @ivelsko, who put up with the never-ending series or meetings and re-writing over each others comments and suggestions on Google Docs! ❤️ you all!
*Thread Reply:* Brilliant! Huge congratulations to the team!
Oops, sorry - apparently that link was a 'author sharing one' that only has 10 download limits 😆 . Could someone send me the PDF if they downloaded it?
Proper link here 😉: https://journals.asm.org/doi/epub/10.1128/msystems.01315-21
*Thread Reply:* Fecking hell I'm disorganised at the moment 🙄🤦♀️:skintone3:
*Thread Reply:* Thanks you!
Interesting looking preprint from @Meriam Guellil @Lucy van Dorp et al!
https://www.biorxiv.org/content/10.1101/2022.01.19.476912v1

From the newly joined @Eric Capo @Marie-Eve Monchamp @Linda Armbrecht!
one for #TeamDir 😉
https://sfamjournals.onlinelibrary.wiley.com/doi/10.1111/1462-2920.15913
Next up on the 2022 preprints! @Pooja Swali et al. With ancient British plague!
https://www.biorxiv.org/content/10.1101/2022.01.26.477195v1

Cautionary tale in ancient metagenomic dietary analysis https://www.biorxiv.org/content/10.1101/2022.01.26.472964v1.abstract?%3Fcollection=

https://www.biorxiv.org/content/10.1101/2022.01.27.477466v1?ct=

https://mbmg.pensoft.net/article/78128/element/8/149541//

Congrats @Meriam Guellil @Marcel Keller and @Freddi Scheib !! Very cool stuff!! https://genomebiology.biomedcentral.com/articles/10.1186/s13059-021-02580-z

https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC8805238/

https://www.nature.com/articles/s41559-021-01652-4
Of interest for the ancient plague community (not ancient DNA though)
Placing ancient DNA sequences into reference phylogenies https://watermark.silverchair.com/msac017.pdf?token=AQECAHi208BE49Ooan9kkhWErcy7Dm3ZL9C[…]yTxKbecBoJeLyWslx_TTOSg2BYFCGTtP-9wzaa8cdpy7DQq-k2YMcHiM
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1009838
https://www.sciencedirect.com/science/article/pii/S147297922200018X?via%3Dihub

Congrats to @Åshild (Ash) on her new paper! (and thank you for the AncientMetagenomeDir Issue too :D)
Ancient TB from pre-contact S. American populations!
https://www.nature.com/articles/s41467-022-28562-8
Our paper "Understanding the microbial biogeography of ancient human dentitions to guide study design and interpretation" is finally out! 😄 https://academic.oup.com/femsmicrobes/article/doi/10.1093/femsmc/xtac006/6541847?login=false
Might be of interest: https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.13608
"A Theoretical Analysis of Taxonomic Binning Accuracy "
Beyond dirty teeth: Integrating dental calculus studies with osteoarchaeological parameters https://www.sciencedirect.com/science/article/pii/S1040618222000726

https://peerj.com/articles/12784/

As the authors are being shy:
https://www.pnas.org/doi/10.1073/pnas.2116722119
Congrats to @aidanva @Gunnar Neumann @Maria Spyrou !
Thanks guys! Very glad it’s out! Big congratulations to Aida, Gunnar and Lyazzat for all their work!
Curious about the kinds of biomedical questions that can be addressed with ancient dental calculus? How much does it reflect oral health? Just how much does community structure change concomitant with human cultural transitions? Check out our new preprint! https://www.biorxiv.org/content/10.1101/2022.04.25.489366v1

We've started to have an uptick in follows on the SPAAM #twitter account - so I would like to make it a bit more active. I thought one way would be to post papers more regularly (e.g. Ancient Metagenomic Paper of the WEek) - would do people think about this paper: https://www.annualreviews.org/doi/abs/10.1146/annurev-phyto-021021-041830
Would that be a good one to share on Twitter?
Ok scheduled! @channel please post new papers you come across here, and we can pick one each week to post on the twitter account 🙂. Bonus points if it's from SPAAM members!
Not a paper, but following our discussion with @Nikolay Oskolkov @Zoé Pochon @Meriam Guellil, some playing with KrakenUniq https://maximeborry.com/post/kraken-uniq/
Congrats @Antonio Fernandez-Guerra
https://elifesciences.org/articles/67667
*Thread Reply:* Congrats indeed Antonio!
Thanks @James Fellows Yates 😀 Check Chiara’s tweet thread for methodological aspects of the paper https://twitter.com/chiaravanni5/status/1531330691337789441
And Matt's for a more biological view https://twitter.com/mschecht_bio/status/1531329448003371009
I talked about it last year in SPAAM3, and now it is published ! sam2lca is now officially released 🙂 https://joss.theoj.org/papers/10.21105/joss.04360 https://github.com/maxibor/sam2lca

Cool, congratulations @Maxime Borry! What is now the output of sam2lca? I remember it was a jason while you were working on outputting a bam with multi-mappers assigned to higher taxonomic levels, is that implemented in this release?
*Thread Reply:* https://sam2lca.readthedocs.io/en/latest/output.html 😉
*Thread Reply:* Awesome, thanks a lot @Maxime Borry, I am going to try it!
New preprint from @Markella Moraitou @Adrian Forsythe and @Katerina Guschanski!
https://www.biorxiv.org/content/10.1101/2022.06.06.494923v1
"Dental calculus metagenomics suggest that ecology, not host phylogeny, shapes the oral microbiome in closely related species" - in ~19th/20th Gorilla dental calculus samples!
Not exactly a paper, but:
Reproducible, scalable, and portable De Novo assembly pipeline... Now with ancient DNA mode courtesy of @Maxime Borry and @Alex Hübner;)
https://www.biorxiv.org/content/10.1101/2021.11.06.467582v3.abstract

I wonder if this would be useful for identifying true 'ancient' calculus, or just caluclus vs envirnomental stuff
(and this was the wrong slack channel, still applies though! )
https://www.nature.com/articles/s41598-022-13269-z
*Thread Reply:* Amazing! Great work Maria, congratulations!
From @Maria Spyrou et al
https://www.nature.com/articles/s42003-022-03527-1
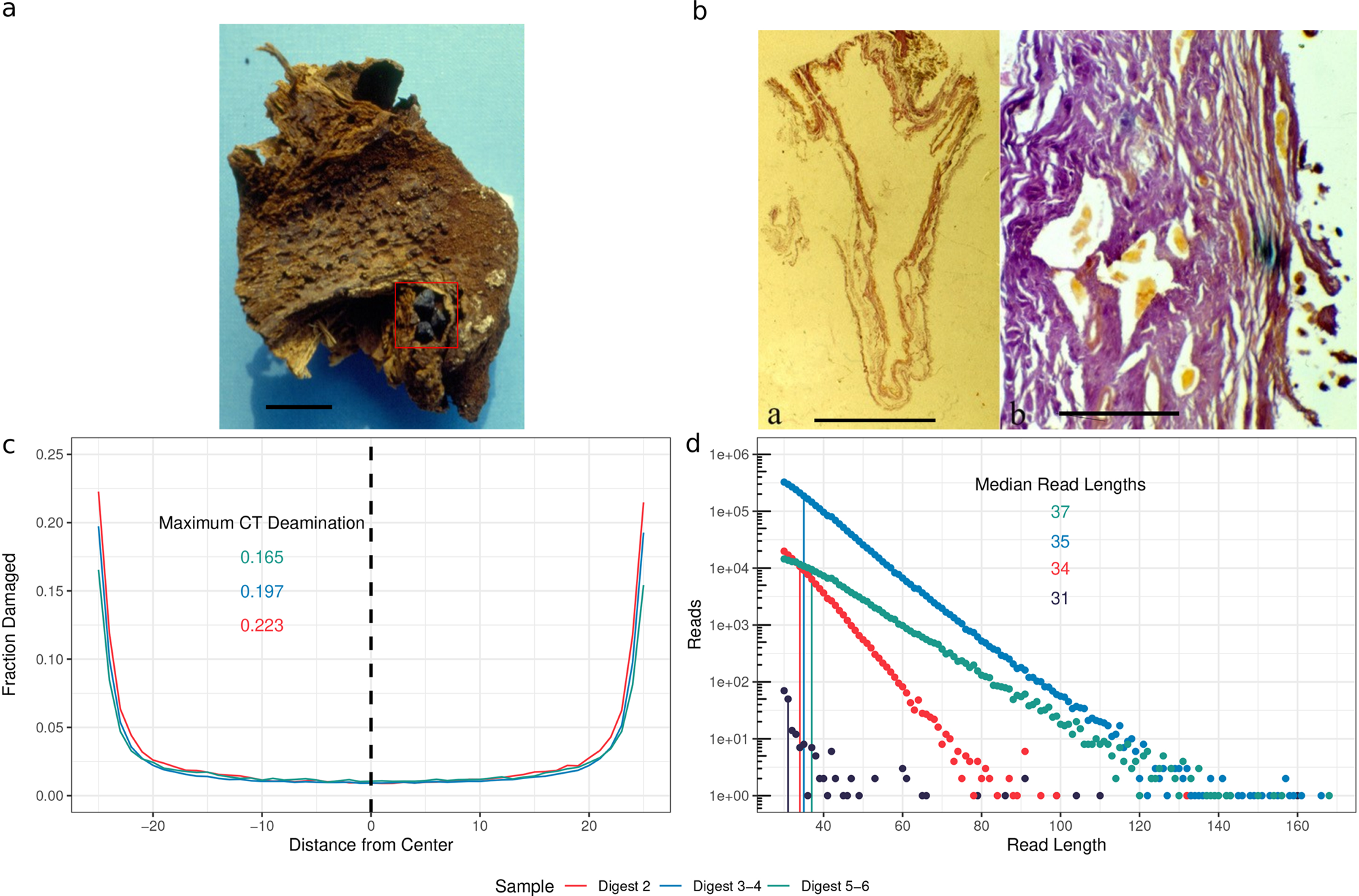
*Thread Reply:* good enough for paper of the week on twitter @aidanva?
*Thread Reply:* i'll schedule t for weds and you to give you time to change your mind
I’ve been a bigger fan of Kraken2 so far (for its lower memory footprint and faster speed), but looks like KrakenUniq now has a nice point for itself ! https://www.biorxiv.org/content/10.1101/2022.06.01.494344v1

*Thread Reply:* That is, if you can build a DB for it
*Thread Reply:* Yes, agree, potentially, a game changer. And a database can be shared. We have a nice collection of large databases for KrakenUniq
https://www.nature.com/articles/s41564-022-01116-w

https://www.biorxiv.org/content/10.1101/2022.06.30.498301v1

nature.com/articles/s41598-022-10690-2
For the #ancientmetagenomedir ?
I don't think so, it doesn't look like they recover genomes
Also it's massively overamplieid (very stacky/duplicated) so I think it's unikely you could get there either sadly
(not ancient, but maybe partially relevant):
thoughts @Antonio Fernandez-Guerra?
yet another method based on consensus taxonomic classification of contigs in a MAG. Always is a good practice to run any of these methods to label potential problems
and of course, the microbial dark matter stuff: https://merenlab.org/2017/06/22/microbial-dark-matter/
*Thread Reply:* Never fails to make me smile
*Thread Reply:* microbial marketing gone bad
using mmseqs2 and their contig taxonomic workflow (https://academic.oup.com/bioinformatics/article/37/18/3029/6178277) you can get a similar output than what MDMcleaner is doing
I like this one https://github.com/dib-lab/charcoal
although in all cases there are strong limitations when using taxonomic information
we will be releasing soon some new methods to clean ancient assemblies and MAGs, will post it here when they are ready for others to use
I haven’t also been working on some sort of pipeline based on a two-step MMSeqs2 taxonomy approach to automatically remove potential wrongly binned and chimeric contigs. We have been using internally successfully for a couple of projects but it still needs more exploration and find tuning, I guess: https://github.com/alexhbnr/automatic_MAG_refinement I would like to know your thoughts on this, too, @Antonio Fernandez-Guerra! I was planning to contact you on this later this summer but will now take the opportunity! 🙂
Cool @Alex Hübner I will have a look and come back to you
Congrats @Gunnar Neumann! “Ancient Yersinia pestis and Salmonella enterica genomes from Bronze Age Crete” https://www.sciencedirect.com/science/article/pii/S0960982222011010?via%3Dihub

And congrats to @Meriam Guellil!
Congrats to @Vilma Perez (or @Vilma Pérez ?)
https://www.mdpi.com/2076-2607/10/8/1623

*Thread Reply:* Do you want the SPAAM twitter to tweet it @Vilma Pérez /@Vilma Perez?
*Thread Reply:* hey @James Fellows Yates I opened slack after many many months, apologies I didn’t your messages before!.
*Thread Reply:* No worries!
From @irinavelsko!
Congrats to @Jessica Hider who is somehow publishing from the future 🤔
https://www.sciencedirect.com/science/article/pii/S1879981722000420?dgcid=coauthor

"Examining pathogen DNA recovery across the remains of a 14th century Italian friar (Blessed Sante) infected with Brucella melitensis"
*Thread Reply:* Thanks @James Fellows Yates :partyparrot: (sorry, I'm slacking off on my slack lol). I'm also confused about my future publishing, very impressive of me lol.
https://www.sciencedirect.com/science/article/pii/S0960982222014671
Not metagenomics but very closely related:
Also including @Åshild (Ash) @Anna F. (And likely a bunch of others sorry, too long author list)

https://www.nature.com/articles/s41596-022-00738-y
I think this is a useful one for many here, mostly those that are just jumping into the ancient metagenomics stuff
https://www.nature.com/articles/s41598-022-20193-9
Another one for the AncientMetagenomeDir
Oh and 16S, but it needs to e shotgun 😉
Also:
*Thread Reply:* How is that possible?! Scientific Reports is a Nature publication, don’t they require you put the data online?
*Thread Reply:* They require you to pay them money 💰
haaha, ok, my bad, I didn’t see all that… I just read the title and very quickly the abstract and asumed it was shotgun
https://www.biorxiv.org/content/10.1101/2022.10.03.510579v1

"aMeta: an accurate and memory-efficient ancient Metagenomic profiling workflow"
From @Zoé Pochon @Nora Bergfeldt @Emrah Kırdök @Tom van der Valk @Nikolay Oskolkov and co!
(I guess you'll need to update your #SPAAM4 title @Nikolay Oskolkov 😉 )
". Here, we propose aMeta, an accurate ancient Metagenomic profiling workflow designed primarily to minimize the amount of false discoveries and computer memory requirements."
*Thread Reply:* Thank you James! I have already let @Shreya know the new title of the talk at SPAAM4 🙂
*Thread Reply:* Thanks! I hope people will find the pipeline useful 😊 (I pushed for a shorter name for it, mea culpa 😅)
*Thread Reply:* @Zoé Pochon i still think MEAD would have been funnier 😅 😉 😘
*Thread Reply:* Great that is already out! I was waiting for it @Nikolay Oskolkov! Looking forward to test it!
*Thread Reply:* Are the DBs already available for downloading?
*Thread Reply:* @Nico Rascovan dude up your twitter game, @Nikolay Oskolkov tweeted that couple of days ago 😉
*Thread Reply:* Thank you @James Fellows Yates and thanks to the co authors @Zoé Pochon @Nora Bergfeldt @Nikolay Oskolkov @Tom van der Valk for this great work!
*Thread Reply:* https://twitter.com/NikolayOskolkov/status/1575009604437561345
*Thread Reply:* Yes @Nico Rascovan, the links are mentioned in the preprint, but I am also going to post them in the "general" channel, hope they will be of interest for many people
*Thread Reply:* hahaha, I barely check twitter…. If it does not send me a notification to my phone, I won’t see it
*Thread Reply:* shakes head
*Thread Reply:* @Nikolay Oskolkov Do you have the full NCBI DB indexed for KrakenUniq? I think Eukaryotic reads are highly underexplored
*Thread Reply:* Thank you @Nico Rascovan for re-twitting! 🙏 The full NCBI NT is available here
KrakenUniq database based on full NCBI NT: <https://doi.org/10.17044/scilifelab.20205504>
*Thread Reply:* @Nico Rascovan this database contains both eukaryotic and prokaryotic references. However, while prokaryotic references are often complete genomes or chromosome level assemblies, the eukaryotic references are usually of a bad quality, so only pieces of reference genomes. This has advantages and disadvantages. The advantage is that the DB is possible to fit to the RAM and that the eukaryotic references can be used as a decoy to attract potential eukaryotic contamination. The disadvantage is that the varying quality of eukaryotic references creates a "database bias", i.e. when one discovers e.g. a wolf in a metagenomic sample just because wolf happened to have a better quality reference (included into NT) compared to the reference of bison that is truly present in the sample
*Thread Reply:* Yes, I totally see the point. But including any known sequence (at least any sequence in the NCBI nt db) would allow reducing a bit the high percentage of unclassified reads you get with KrakenUniq. Then, as you mention and shown already before, there are downstream analyses that can be done to further authenticate, characterize or understand what do certain hits mean. No kraken results should be taken for granted without some controls.
Oh man, you guuys are on a roll https://www.nature.com/articles/s41467-022-33494-4#Sec11
Congrats to @Linda Armbrecht and team!
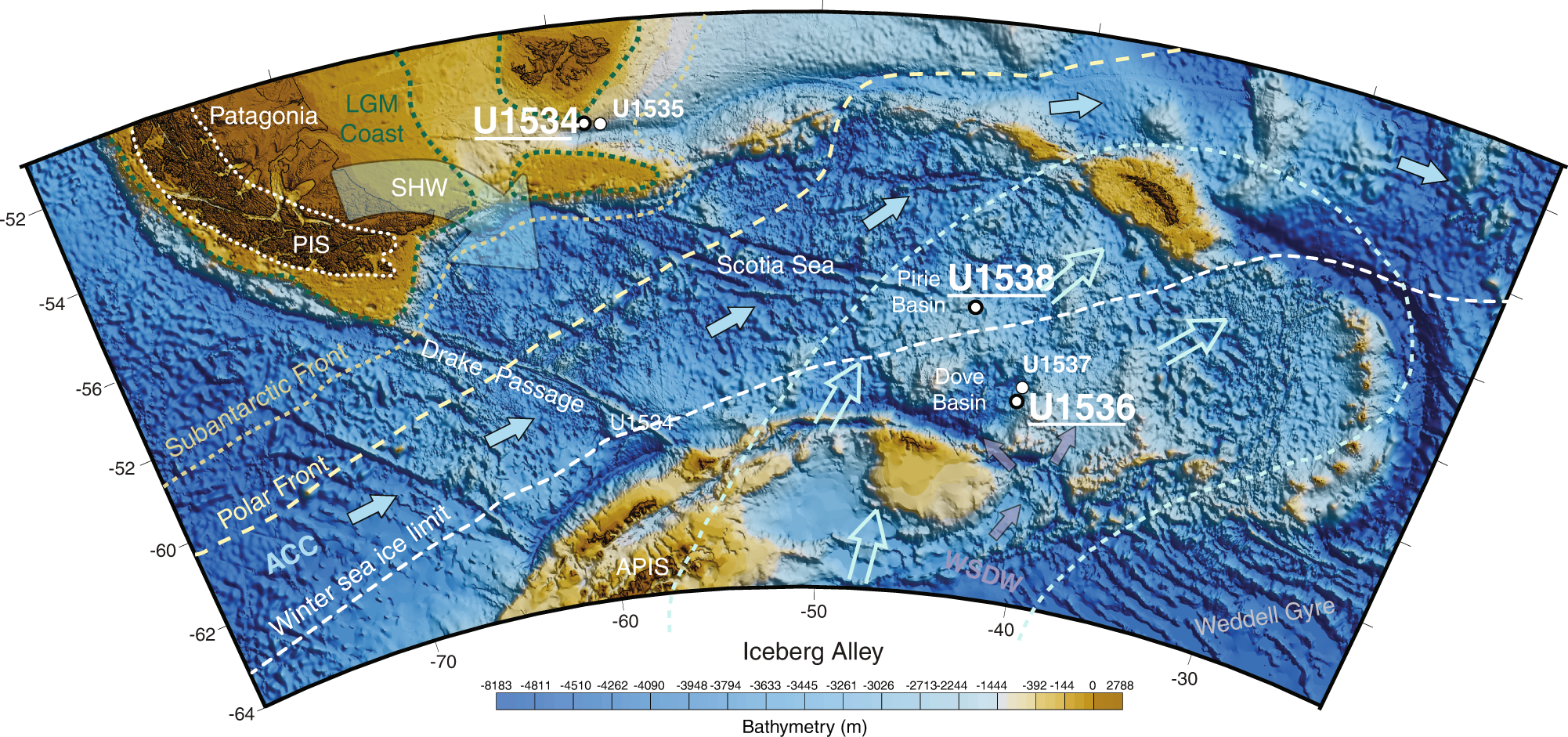
And @Rita M Austin https://www.nature.com/articles/s42003-022-03890-z
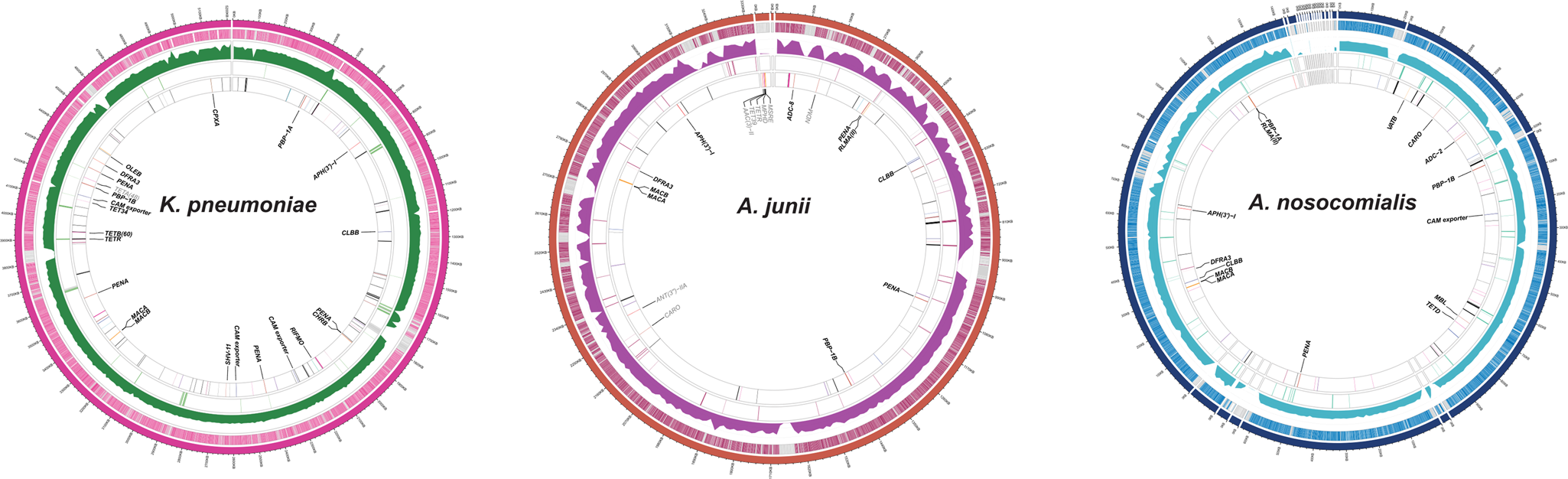
https://www.nature.com/articles/s41586-022-05349-x Cool paper out by SPAAM4 career panelist Jennifer Klunk!
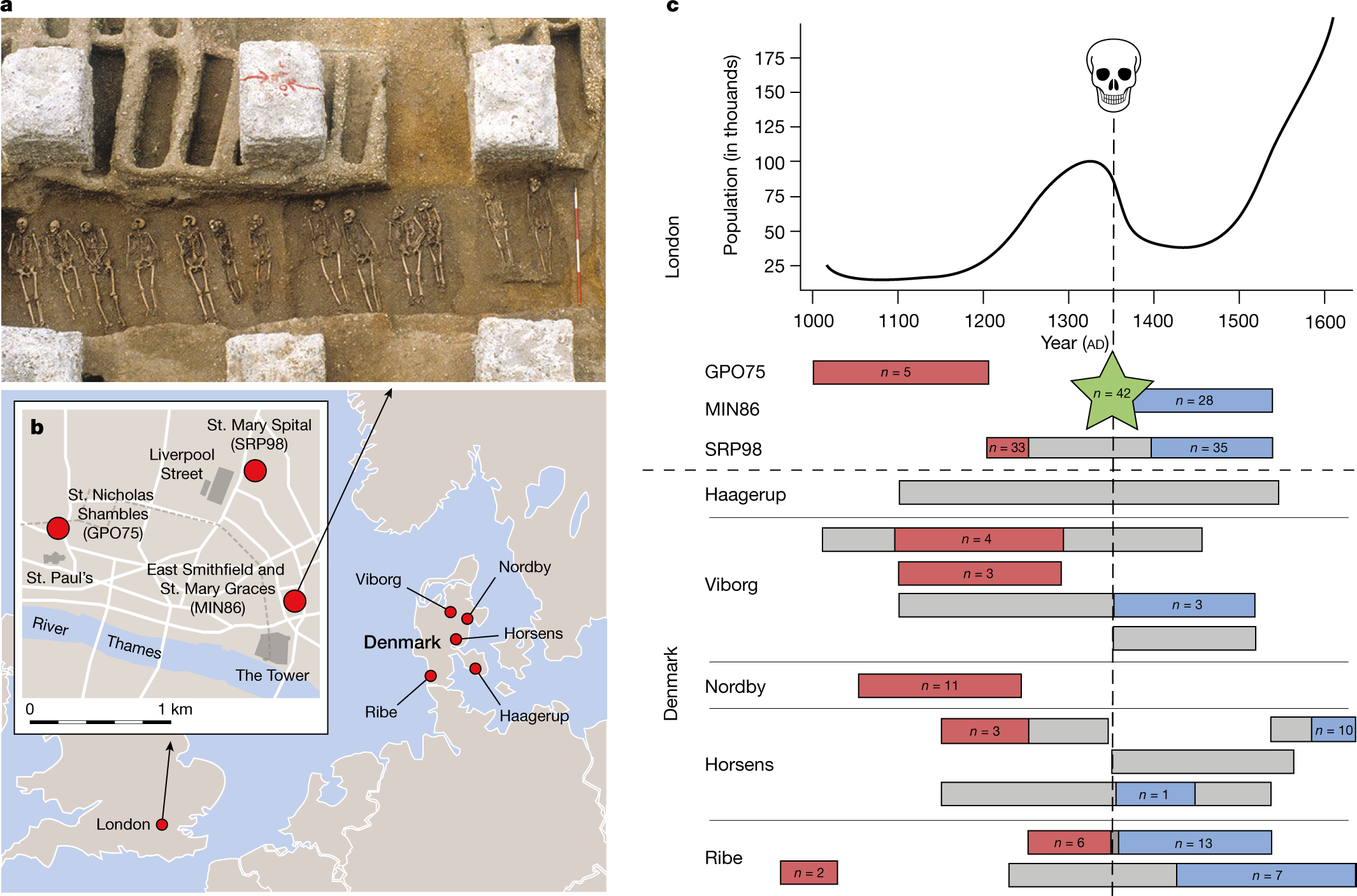
Not metagenomics, but possibly of interest to those working on eDNA metacoding data: https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.13356
Paper and blog post about FAIR principles for research software:
Nature
Research Data at Springer Nature

https://www.nature.com/articles/s42003-022-04190-2
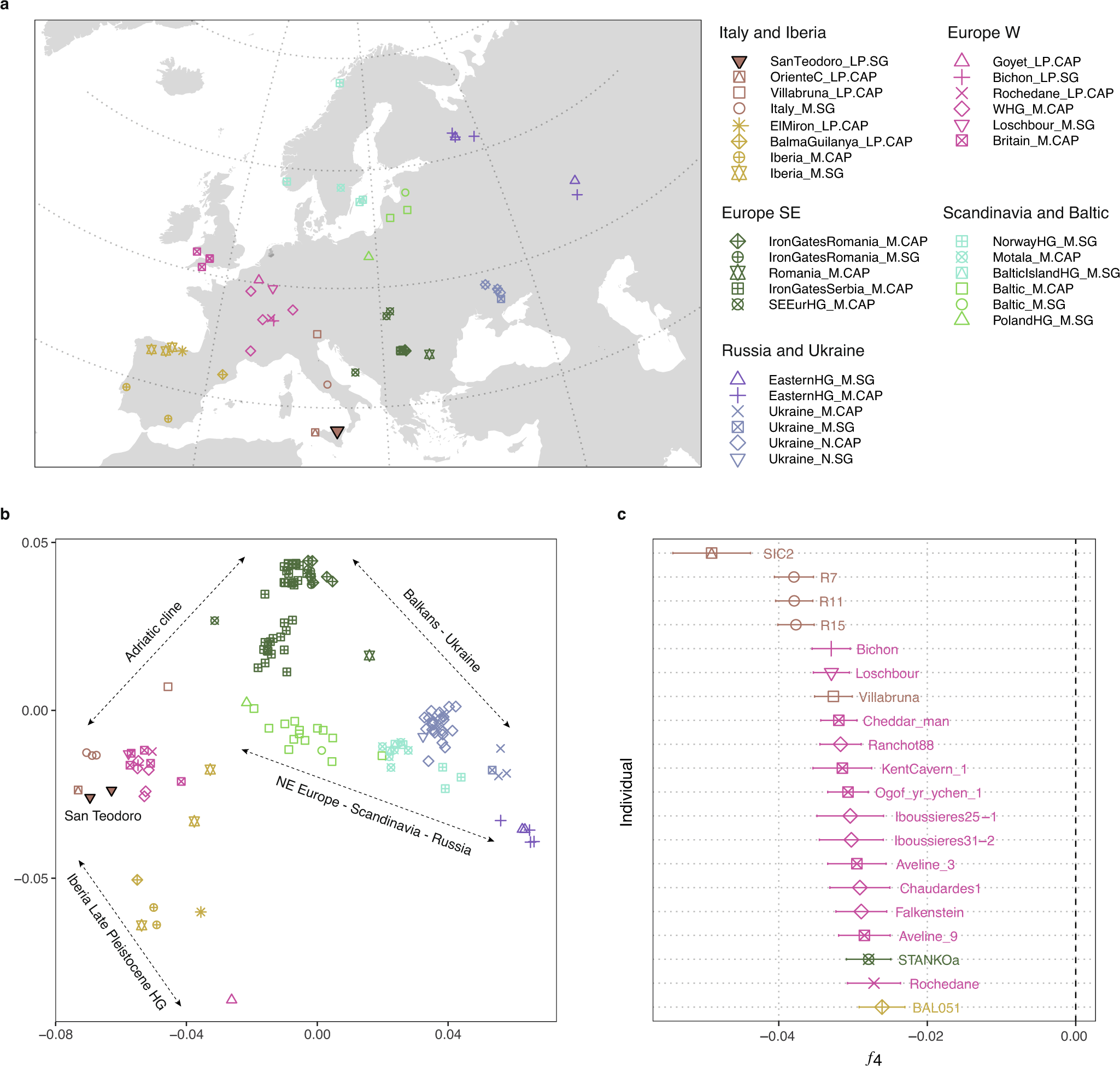
*Thread Reply:* I think this is a paper that worth discussing in the SPAAM community, as they implemented some new strategies to analyze ancient microbiome data, that have not been used before in the field.
*Thread Reply:* Maybe @Andrea quagliariello can make a summary about it for the rest of the community, I think it will be very useful for others
*Thread Reply:* #spaamtisch?
*Thread Reply:* (@Maria Lopopolo)
*Thread Reply:* I can definitely propose it to the community among the topics as broad discussion although this is more JC material. But well in our first #spaamtisch we'll see what people think the channel should be dedicated to ;)
*Thread Reply:* Thank you @Nico Rascovan yes I would be delighted to do that for the community!!! But you have to wait some days because, unfortunately, I am sick at home with flu 🥲. Thus, instead of going out with my collegues to celebrate it, I am at home with the blanket and the hot water bottle:mask_parrot:🤣. I promise I will come back soon🙏💪
*Thread Reply:* Get well soon!
*Thread Reply:* OK it would be coolto have an open discussion, I already have questions!
*Thread Reply:* Great study @Andrea quagliariello congratulations!
Ancient oral microbiomes support gradual Neolithic dietary shifts towards agriculture
Congrats to @Andrea quagliariello!
*Thread Reply:* Thank you @James Fellows Yates!!!🙂
*Thread Reply:* Btw ❤️ for citing protocols.io protocols 😄
*Thread Reply:* 😄of course!!! 🙂
It is a very nice article! 🎉👏
You can tell it's coming to the end of the year 😆
https://www.biorxiv.org/content/10.1101/2022.01.27.477466v1.full

From @Diana Ivette Cruz Dávalos @Miriam Bravo @Samuel Neuenschwander @Viridiana Villa and co!
Jacob Haffner here at LMAMR published this awesome paper on the human metabolome!
Untargeted Fecal Metabolomic Analyses across an Industrialization Gradient Reveal Shared Metabolites and Impact of Industrialization on Fecal Microbiome-Metabolome Interactions https://journals.asm.org/doi/full/10.1128/msystems.00710-22
https://academic.oup.com/nargab/article/4/4/lqac090/6855701
*Thread Reply:* @Alex Hübner @irinavelsko
> *Comparison of functional classification systems * > In microbiome analysis, functional profiling is based on assigning reads or contigs to terms or nodes in a functional classification system. There are a number of large, general-purpose functional classifications that are in use, such as eggNOG, KEGG, InterPro and SEED. Smaller, special-purpose classifications include CARD, EC, MetaCyc and VFDB. Here, we compare the different classifications in terms of their overlap, redundancy, structure and assignment rates. We also provide mappings between main concepts in different classifications. For the large classifications, we find that eggNOG performs the best with respect to sequence redundancy and structure, SEED has the cleanest hierarchy, whereas KEGG and InterPro:BP might be more informative for medical applications. We illustrate the practical assignment rates for different classifications using a number of metagenomic samples
https://www.nature.com/articles/s41586-022-05453-y
2-Million years old sedaDNA! Impressive! Congrats @Antonio Fernandez-Guerra that participated in the work and presented some exciting results of the study in our institute last week!
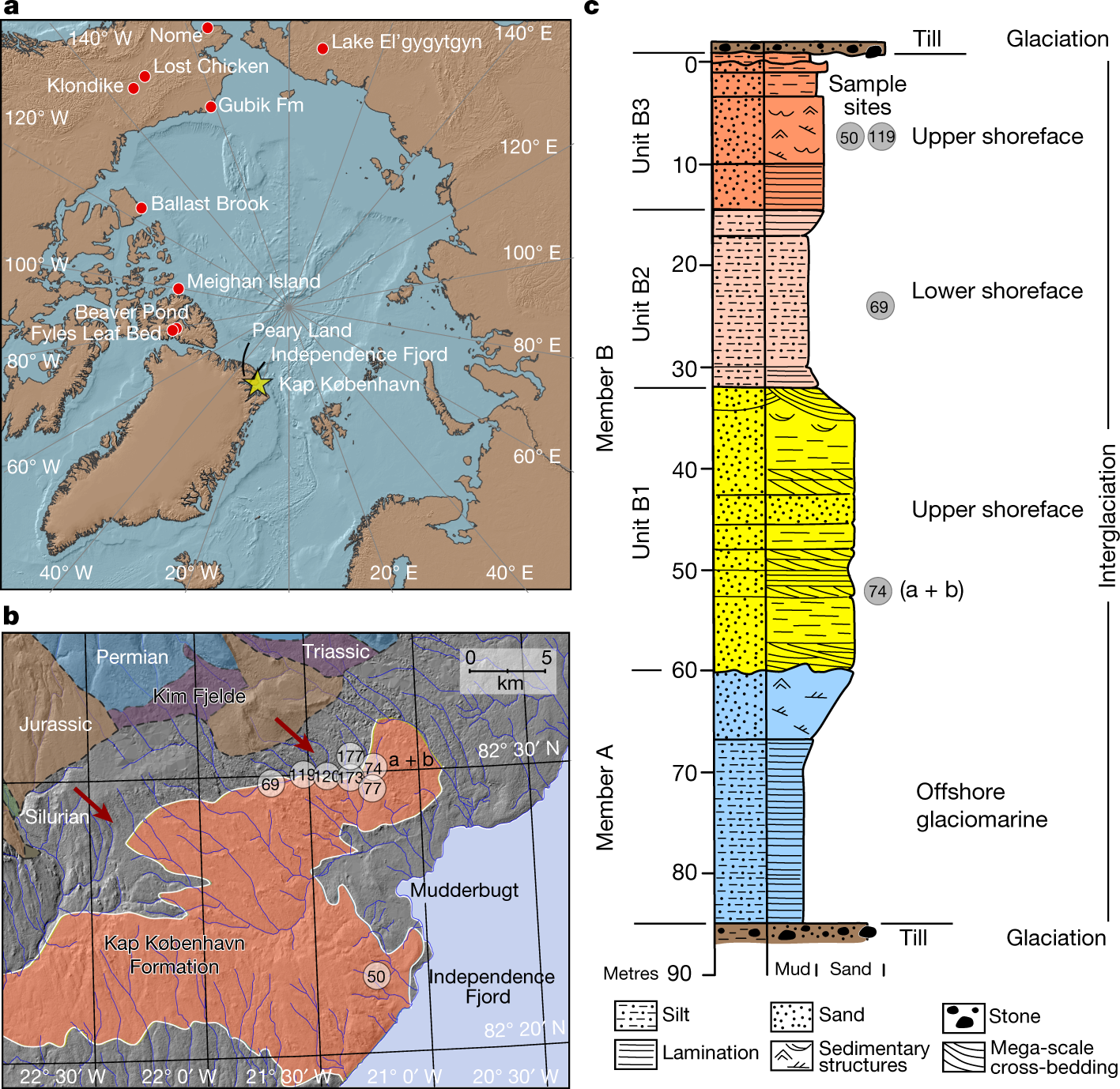
*Thread Reply:* the race has begun 🙂
*Thread Reply:* Race to finding the oldest DNA again ? Back to amber ?
And also to @Markella Moraitou @Adrian Forsythe @Jaelle Brealey @Katerina Guschanski
@Markella Moraitou has joined the channel
@Adrian Forsythe has joined the channel
MEGAN-Server... Is back...?
https://www.biorxiv.org/content/10.1101/2022.12.05.518498v1?rss=1

*Thread Reply:* Was it ever gone? Haven’t used it since 2017 I think
*Thread Reply:* As in it stopped being developed
Congratulations @Andrea quagliariello !!
But my institution doesn't pay for access to the journal so I can't read it 😭😭
(hope that's OK @Andrea quagliariello!)
*Thread Reply:* Absolutely yes!! thank you @James Fellows Yates Also because I requested to make it open access
https://doi.org/10.1186/s13059-022-02806-8

*Thread Reply:* Congrats @Joanna H. Bonczarowska!
*Thread Reply:* Would you be interesting in adding the genomes to AncientMetagenomeDir? https://github.com/SPAAM-community/AncientMetagenomeDir
Anyone who adds metadata will be included on the next publication 🙂
*Thread Reply:* As you've got only 7 genomes it would probably take only about 30m for you
*Thread Reply:* (sorry to evangelise 😆 )
*Thread Reply:* @James Fellows Yates only the pathogen-positive metagenomes? I will check it out :)
*Thread Reply:* And also if you're on Twitter - have you made a twitter thread or something like that about he paper? We can retweet from the SPAAM account
*Thread Reply:* > only the pathogen-positive metagenomes? I will check it out 🙂 Not even, just the genomes themselves
*Thread Reply:* Unless you've analysed calculus or something (but I don't see that mentioned)
*Thread Reply:* Unfortunately, I do not have twitter, not sure if any of my co-authors do.
*Thread Reply:* Ok, I can tweet juts the paper 🙂
*Thread Reply:* Thank you
*Thread Reply:* https://twitter.com/spaam_community/status/1602580490736787457
*Thread Reply:* I hopes thats Ok
*Thread Reply:* That’s great thank you :)
Hi All, can anyone suggest papers that have looked at the causes of DNA degradation in ancient samples? Particularly any reviews?
*Thread Reply:* https://royalsocietypublishing.org/doi/10.1098/rspb.2004.2813 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1802572/ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC96698/ The first chapter of the book has a good table on damage types


The ‘other’ Dabney et al. 2013 paper: https://cshperspectives.cshlp.org/content/5/7/a012567.full
Interesting thread and papers about taxonomic classification/profiling: https://twitter.com/SilasKieser/status/1613868658811600896
 }
Dr. Silas Kieser @silask@mstdn.science
(https://twitter.com/SilasKieser/status/1613868658811600896)
}
Dr. Silas Kieser @silask@mstdn.science
(https://twitter.com/SilasKieser/status/1613868658811600896)
https://animalmicrobiome.biomedcentral.com/articles/10.1186/s42523-022-00207-7

this is my take on it https://github.com/genomewalker/bam-filter
here it provides abundance estimates and applies multiple filters beyond breadth
in any case having accurate abundance estimates of ancient metagenomic data is extremely difficult
This is based on BAM files and is heavily coupled with our DBs and our metaDMG tool (https://www.biorxiv.org/content/10.1101/2022.12.06.519264v1)

filterBAM applies different filters beyond the traditional ones like positional entropy of the reads and report a few abundance estimates
I strongly recommend you to take a look at @Antonio Fernandez-Guerra take on all this (i.e., his pre-prints and github). He is applying it to the analyses of ancient microbial genomes from sediments, he recently explained us many of them in more detail in a visit to our lab, and I really loved it! @Antonio Fernandez-Guerra, you should give a seminar for the SPAAM community! Shouldn’t we have one monthly seminar/Lecture in SPAAM?
*Thread Reply:* If you volunteer to set it up/run it @Nico Rascovan 😉
*Thread Reply:* I’ll talk with the people of my team to organize it, all right! It’s just setting up one seminar per month, not big deal I hope ;)
*Thread Reply:* Note that @Maria Lopopolo is already running the #spaamtisch!
*Thread Reply:* Could also consider integrating the two
*Thread Reply:* Evening seminar style thing
*Thread Reply:* (sponteaenous ly, )
*Thread Reply:* > I am aware… yes Needed a way to ping her 😉
New mapping pipeline (in snakemake 👀) https://pubmed.ncbi.nlm.nih.gov/36637197
Congratulations to @Samuel Neuenschwander @Diana Ivette Cruz Dávalos @Davide Bozzi!
Ancient pathogens provide a window into health and well-being https://www.pnas.org/doi/10.1073/pnas.2209476119
Congrats to @Katherine Eaton!
https://www.nature.com/articles/s42003-022-04394-6
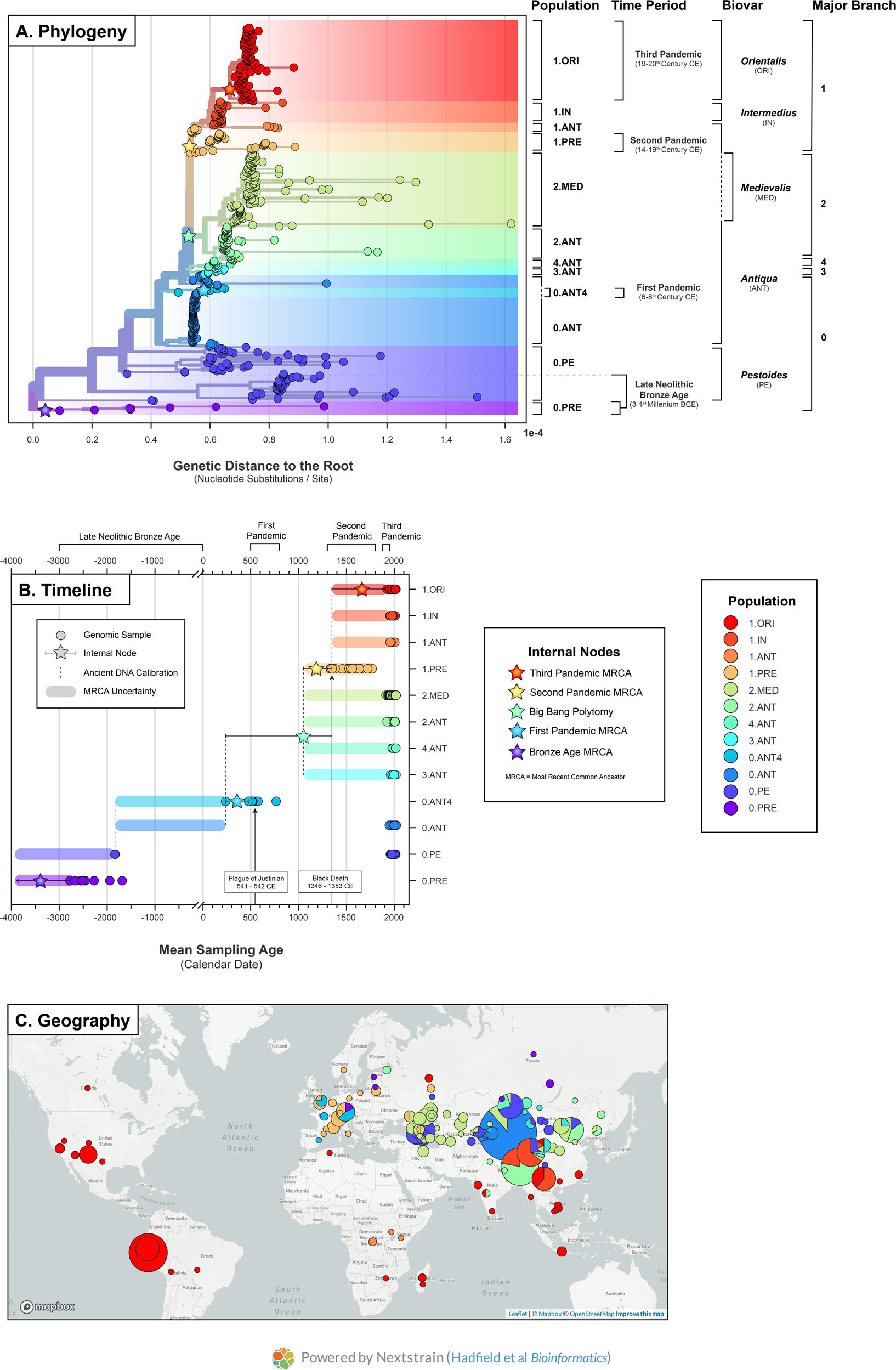
*Thread Reply:* From the people who brought MixOmixs @irinavelsko
https://www.frontiersin.org/articles/10.3389/fevo.2022.888421/full
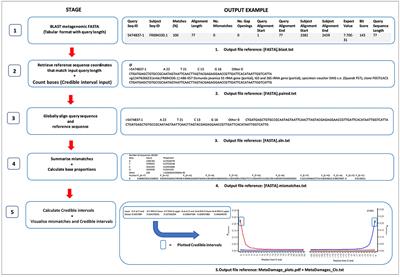
From Rosie Everett and ex-SPAAM Becky Cribdon in Warwick
TL;DR Damage profiles from BLAST output
With some extra stats about confidence in the profile
@Camila Duitama @Nico Rascovan
You post a preprint on an ancient metagenomic source tracking tool and you don't post it here 😱😱😱?!?
Congrats though!
https://www.biorxiv.org/content/10.1101/2023.01.26.525439v1?rss=1

Hi @James Fellows Yates and everyone! We had to post the preprint at the last minute, as we are preparing another submission 🙂. We are looking forward to hearing your comments, we hope decOM will help the aDNA community with your analysis 🧹🦷.
https://www.biorxiv.org/content/10.1101/2023.01.24.524926v1
Preprint of a review from @Abby Gancz as well!
https://f1000research.com/articles/12-109

Congrats to @Mohamed Sarhan @Christina Wurst!
https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-022-01509-7

*Thread Reply:* @Mohamed Sarhan interested in adding your data to #ancientmetagenomedir?
*Thread Reply:* https://github.com/SPAAM-community/AncientMetagenomeDir/issues/1045
*Thread Reply:* Sure!
And @Samuel Neuenschwander and @Diana Ivette Cruz Dávalos
https://academic.oup.com/bioinformatics/article/39/1/btad028/6986969?login=false
Also from @Mohamed Sarhan’s paper
:partyparrot: :partyparrot: :partyparrot: :partyparrot: :partyparrot:
*Thread Reply:* Thanks again for your comments! They helped a lot with the peer-reviewing 😊 @James Fellows Yates @Nikolay Oskolkov @irinavelsko
Not aDNA per-se, but could be very relevant given commonly fragmented assemblies, from @Raphael Eisenhofer!
https://www.nature.com/articles/s43705-023-00221-z

*Thread Reply:* (why, are you planning to help add it to #ancientmetagenomedir ? 😬 )
*Thread Reply:* perhaps, but not for at least a few weeks -- the first author could as well 😉
*Thread Reply:* hahaha, teasing, no worries 😉
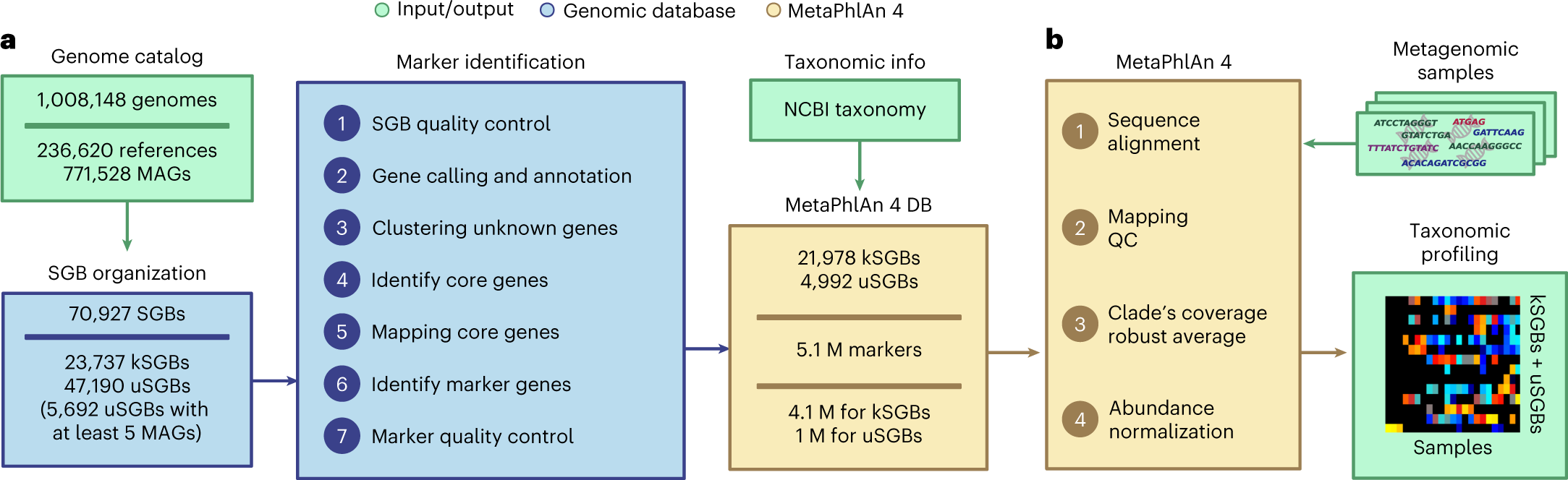
From @Katherine Eaton and co!
MetaPhlAn4: https://www.nature.com/articles/s41587-023-01688-w
https://www.nature.com/articles/s42003-023-04582-y
Really nice recommendations here on when to use Kraken2 vs MetaPhlAn3, and how to construct your database and/or which settings to use: https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000949
Note a paper, but close enough (warning self PR):
https://twitter.com/jfy133/status/1635277477575811081
 }
James Fellows Yates | @jfy133@genomic.social
(https://twitter.com/jfy133/status/1635277477575811081)
}
James Fellows Yates | @jfy133@genomic.social
(https://twitter.com/jfy133/status/1635277477575811081)

git (not aDNA specific but may add aDNA speicfic steps in the future)
https://www.nature.com/articles/s41587-023-01696-w
“Ancient DNA and Forensic Microbiology” is now fully online and open for submissions!
*Thread Reply:* Do you have a link?
*Thread Reply:* Is this a journal or special issue of one?
*Thread Reply:* It is a new section of Frontiers in Microbiology
*Thread Reply:* https://www.frontiersin.org/journals/microbiology/sections/ancient-dna-and-forensic-microbiology

https://academic.oup.com/bib/article/24/2/bbad092/7077155
I wonder how this would perform on ancient DNA datasets… if it works well, it could be a good fast screening option..
De Sanctis, B. (2022). Phylogenetic inference using ancient environmental DNA (Doctoral thesis). https://doi.org/10.17863/CAM.95210
https://aspace.repository.cam.ac.uk/handle/1810/347793
https://onlinelibrary.wiley.com/doi/10.1002/ajpa.24735
From Tanvi Honap, @Rita M Austin and co
*Thread Reply:* To be added in the AncientMetagenomeDir, right?
*Thread Reply:* No public data unfortunately:(
*Thread Reply:* As far as o can tell
*Thread Reply:* Where did you find that?!
*Thread Reply:* I looked through the paper twice yday?!
*Thread Reply:* It's hidden if you're on the website, click the Open Research dropdown at the end of the page
*Thread Reply:* Thanks @irinavelsko I guess that's a lesson in not looking via phone
*Thread Reply:* It's not at all obvious, it took me a while to find it
*Thread Reply:* It's terrible isn't it?!?
https://www.nature.com/articles/s42003-023-04723-3
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10061425/

*Thread Reply:* Love the acronym 😆
From @Alex Hübner, @Anan Ibrahim and co!
https://www.science.org/doi/10.1126/science.adf5300
*Thread Reply:* Although this brings a dilemma now @Alex Hübner, how shall we integrate the new libraries into AncientMetagenomeDir 🤔
*Thread Reply:* Congratulations guys 🎉🎉🎉
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10150486/

Hi y'all, is there someone here who might be able to provide access to this article? Warinner, Christina (2022): An Archaeology of Microbes. In Journal of Anthropological Research 78 (4), pp. 420–458. DOI: https://www.journals.uchicago.edu/doi/10.1086/721976 (Please delete if this post is out of place), thank you in advance 🙂
*Thread Reply:* @Tina Warinner?
*Thread Reply:* But this is the right place @Freya Steinhagen 🙂
*Thread Reply:* I happened to have just read this article!
*Thread Reply:* Wow, that was fast! Thanks a lot <3
*Thread Reply:* Sorry - just seeing this now. Thanks for sending it @Yuti Gao!
From @Bjorn Bartholdy https://www.biorxiv.org/content/10.1101/2023.05.23.541904v1.full
Congrats to @Pooja Swali et al!
https://www.nature.com/articles/s41467-023-38393-w
Hello everyone, I'm excited to share my first paper on ancient metagenomics.
Our study shows that not only ancient bacteria but also their enemies, bacteriophages, can be reconstructed de novo from ancient metagenomes. Despite the perception of phages as highly variable due to mutations, recombinations, and gene transfers, we found that sometimes they can exhibit significant stability over time.
https://www.biorxiv.org/content/10.1101/2023.06.01.543182v1
*Thread Reply:* Nice figures btw!
*Thread Reply:* Thanks, prepared by #AndrzejZielezinski 😉
Congratulations!
(Congrats to @Antonio Fernandez-Guerra, finally ;) )

*Thread Reply:* Thanks @James Fellows Yates Soon the second part with the 2Myr MAGs 😃
https://www.nature.com/articles/s41559-023-02056-2
https://www.biorxiv.org/content/10.1101/2023.06.13.544823v1

Nice technology feature! https://www.nature.com/articles/d41586-023-02154-y Props to @Pooja Swali for advertising <#C02DCKJ54JX|no-stupid-questions> and #ancientmetagenomedir 😁

https://www.biorxiv.org/content/10.1101/2023.07.17.549303v1?rss=1

https://www.biorxiv.org/content/10.1101/2023.07.28.550993v1

Some interesting technical insights there on what can happen when you don't remove human DNA
THE DAY IS HERE !!!! https://f1000research.com/articles/12-926/v1 🔥:partyparrot::spaam:

this is not really SPAAM material, but I’m wondering what people here think about this paper: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1010798 The only evidence for the age of the nematode is radiocarbon dating of the plant material of a rodent burrow where the nematodes were found. To me this is not sufficient evidence for these extraordinary claims.
*Thread Reply:* Agreed, I feel there is a lot lacking here

https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10457426/

Apparnetly they beieve they will also be able to identify the original source of 'ancient' MAGs?
Bacterial pangenome annotation: http://m.genome.cshlp.org/content/early/2023/08/24/gr.277733.123
https://www.nature.com/articles/s41467-023-41174-0
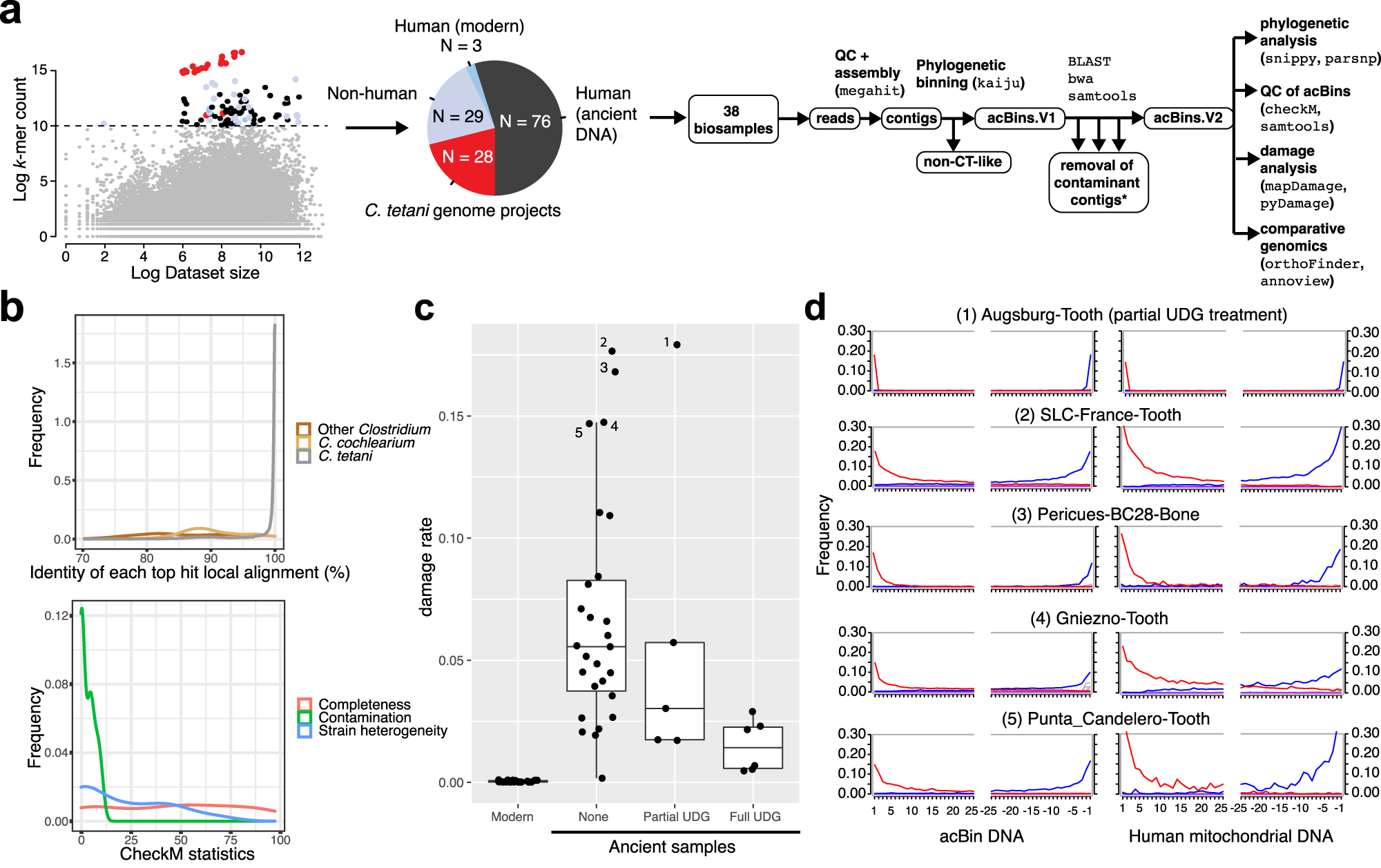
*Thread Reply:* Damaged Clostridium tetani is super common to find in ancient human (and non-human) remains, but is it really an ancient pathogen, i.e. have something to do with ancient tetanus disease? Need to read the paper first 🙂
*Thread Reply:* No it's not an ancient pathologeb
*Thread Reply:* Abs they don't make their claim
*Thread Reply:* Clostridium is general necrobiome, so it's not tetani Most likely
*Thread Reply:* Aida did a masters thesis on it
*Thread Reply:* I just read the abstract and it says "Thus, our ancient DNA analysis identifies DNA from neurotoxigenic C. tetani in archaeological human samples, and a novel variant of TeNT that can cause disease in mammals."
*Thread Reply:* The tree is fucking awful so m'identifico is a mess, as well as tetani being massively over represented
*Thread Reply:* Yes so they get the toxin that is normally right in tetani, and they tested it(which is the cool thing)
*Thread Reply:* @James Fellows Yates why do you call the tree awful? Choice of genomes to include, lack of support values, ... ?
*Thread Reply:* Sorry, meant the general taxonomy not the tree (and I'm not referring to the tree in the paper!)
The clostridium taxonomy and the naming system is purely based on physical characteristics and random tests but has no correspondence with the actusl genomes
*Thread Reply:* @aidanva can explain more
*Thread Reply:* Sorry to be clear!
*Thread Reply:* The is not necessarily bad!!!
*Thread Reply:* I've not read it closely
*Thread Reply:* But just remember the genus clostridium is a major part of the necrobiome, so it could still be ancient just most likely colonised immediately after death rather than being 'endogenous' to the invidual
*Thread Reply:* Just read the paper first, I believe the authors have been careful in waht they say their results are
*Thread Reply:* Didn't intend to misrepresent what you wrote out-loud. Just wanted to understand!
The premortem/postmortem point is well made and one we think about. A cursed taxonomy, I didn't consider!
Congrats to @Nicola Vogel!!
https://besjournals.onlinelibrary.wiley.com/doi/10.1111/2041-210X.14214
Congrats to @Camila Duitama (I think?!)
I'm confused why I've only got this notification a month later... And I can't find a html version of it... but still!!
*Thread Reply:* Gracias!
Lots of food for thought here (ping @Aleksandra Laura Pach @Camila Duitama @irinavelsko @Nikolay Oskolkov)
https://www.mdpi.com/2076-2607/11/10/2478
One interesting quote:
> Another profiler KrakenUniq, an extension of Kraken, works on exact k-mer matching based on the HyperLogLog algorithm. KrakenUniq performed comparably to Kraken2 at the genus level but not so at the species level, establishing Kraken2 as a better profiler for ancient metagenome profiling. Furthermore, the database requirements for Kraken2 are less than those of KrakenUniq

(Although a bit confused by this statement
> MetaPhlAn4 uses nucleotide BLAST (blastn) with a default E-value threshold of 1 × 10−6 to align reads to marker genes. > I'm pretty sure MP** uses bowtie2?
(but I've not read the paper in depth)
Ah and the first author is here - congrats @Vaidehi Pusadkar!!!
Maybe they mixed version 1 and version 4
Hmmm, looking at figures 1-4 I have difficulty seeing any differences in performance, @James Fellows Yates am I misunderstanding them?
*Thread Reply:* It's rather small yes the differences.
As we discussed at spaam5 databases are the bigger influenc/problem
But I think the point about memory usage in KU vs K2 is the more important but
*Thread Reply:* Also, in addition to the database factor, how you filter Kraken2 and KrakenUniq outputs matters a lot
*Thread Reply:* Yes definitely, I honestly think we need to move on from which classifier to which parameters
*Thread Reply:* If anyone wants a project idea ;)
*Thread Reply:* I know a pipeline that can help with that 😬
*Thread Reply:* As important the filtering is, the parameters to build the db will have a huge impact on it. For example in k2, If you track the kmers for the different taxonomic assignments in dbs built with different kmer/minimizer lengths and minimizer spaces you will have a surprise in how the same read is assigned across the parameter space. Specially since the default db settings in k2 are not suitable for many of our datasets where the modal read lengths are fairly close to the default k2 k-mer length
*Thread Reply:* Maybe not the most popular opinion, but I don’t think current k-mer based methods are suitable for our type of data.
*Thread Reply:* Ha-ha, other approaches (alignment and de-novo) have own difficulties 🙂
*Thread Reply:* We need k-mer methods developed for aDNA
*Thread Reply:* Not adopting methods not designed for our type of data
*Thread Reply:* Also we have very interesting new data structures that are very interesting for this problem, for example https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-02971-4

*Thread Reply:* That have very interesting properties
*Thread Reply:* We are working on some solutions as it is very needed
*Thread Reply:* What aren't you working on @Antonio Fernandez-Guerra 😉?
Maybe we need an emoji reaction for you. Someone asks a question and you just have to react on the post with an emoji and a bot will auto-post 'We are working on that' 😉
*Thread Reply:* > Maybe not the most popular opinion, but I don’t think current k-mer based methods are suitable for our type of data. I don't think that's an unpopular opinion! That's why people keep trying to use alignment stuff like MALT... they fall back to kmer stuff because of computational resource requirements ;)
*Thread Reply:* In our case we need to find ways to deal with billions of reads
*Thread Reply:* and where we only end up using a small proportion
*Thread Reply:* If you're interested in bloom filters (or variants): have you tried ganon (https://github.com/pirovc/ganon)?
*Thread Reply:* yes, not working for our purpose
*Thread Reply:* Does that perform better than k2?
*Thread Reply:* What is your purpose in this case?
*Thread Reply:* i.e no mismatches allowed
*Thread Reply:* in the means that mismatches will not reduce the kmer you can generate
*Thread Reply:* from ganon’s paper https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7355301/#E2

*Thread Reply:* I see. Have you considered asking the developer? He's extremely responsive, and he might be willing to add options to make it more amenable
*Thread Reply:* also it doesn’t have a damage model included
*Thread Reply:* ganon is very good
*Thread Reply:* Good to know if it performs better than e.g. k2 still! But again might be worht speaking to the developer, might be good to integrate into that rather than reinventing the wheel
*Thread Reply:* it is not reinventing the wheel, the project is within another project looking at new data structures
*Thread Reply:* that can index larger datasets
*Thread Reply:* another problem we have
*Thread Reply:* also is not a classifier
*Thread Reply:* what we are after
*Thread Reply:* > Good to know if it performs better than e.g. k2 still! But again might be worht speaking to the developer, might be good to integrate into that rather than reinventing the wheel Yes, I don’t know why is not more used
*Thread Reply:* Age and Advertising 😆 😬
*Thread Reply:* I guess, but it touches all the good spots
*Thread Reply:* I've already added it to taxprofiler so I hope it'll tickles peoples interest
*Thread Reply:* I think ganon also did well in CAMI/LEMMI
*Thread Reply:* Guys, does anyone see a difference in F1-score in Figure 2 between ganon and K2?
*Thread Reply:* is the advantage of ganon in just speed, i.e. not accuracy?
*Thread Reply:* (I think we are talking outside of the paper now I think)
*Thread Reply:* ()Ganon isn't tested in the paper)
*Thread Reply:* (whcih is @Antonio Fernandez-Guerra’s point that it's sad people arne't trying it out)
*Thread Reply:* Sorry, I meant in the Ganon paper
*Thread Reply:* For my taste, beside technical details, this is what makes ganon great: • multiple taxonomy integration (NCBI and GTDB) with lowest common ancestor (LCA) • read reassignment EM algorithm for multi-matching reads • hierarchical use of multiple databases • taxonomic and sequence abundance reports with genome size correction
*Thread Reply:* (I just copied from ganon githubs)
*Thread Reply:* > is the advantage of ganon in just speed, i.e. not accuracy? @Nikolay Oskolkov i usually don’t trust so much the benchmarks in the papers as it is very dataset dependent, and most of the time all methods perform fairly bad in our datasets 😅
*Thread Reply:* That's why we need a proper aDNA classifer benchmarking that is outside the racken and MetaPhlan families 😆
*Thread Reply:* And then we steal whatever fancy model Antonio and co are developing and add it to a classifier 🤣
*Thread Reply:* That performs best
*Thread Reply:* and there are many other problems. Check B of this figure. We didn’t find any other viruses beyond caudoviricites using nucleotides based methods. And we had to switch to amino acids
*Thread Reply:* this is a extreme case as they are deep time samples, but shows the complexity and that can not be reduced to a single approach
*Thread Reply:* and we use quite comprehensive DBs for our profilings. For this pre-print we used:
*Thread Reply:* Perhaps you guys at @Antonio Fernandez-Guerra’s lab have a reliable enough dataset that can be used for benchmarking purposes ? (Thinking of the <#C05SQA7RA81|classifier-committee> )
*Thread Reply:* @Camila Duitama we have our internal datasets that we know them very well and we use them as benchmark, but they are not public yet. What I do for each analysis is to create a synthetic dataset based on the empirical data using https://github.com/genomewalker/aMGSIM. For example, to identify the best parameters for bowtie2 in the Kap K preprint: ```Best search parameters estimation To determine the optimal parameters for different searches, we utilised a workflow (https://github.com/aMG-tk/aMGSIM-smk) that combined the aMAW and aMGSIM (https://github.com/aMG-tk/aMGSIM) tools to generate synthetic metagenomes. We used ten samples from the Kap Kobenhavn formation (Supplementary Table 6) to model the community composition (bacteria, archaea, and viruses), the fragment length distribution, and the damage patterns for each reference. Each synthetic metagenome contained a maximum of 1000 damaged references (determined using Bayesian damage estimates), 500 non-damaged references, and 10 million reads. We then annotated these synthetic metagenomes using the aMAW taxonomy module, exploring different bowtie2 parameters (-N, -k [100, 250, 500, 750, 1000]), read ANI filtering thresholds (92%, 93%, 94%, 95%, 96%), and breadth filtering. We evaluated the sensitivity and specificity of the taxonomic profiling, the abundance estimations, and the damage estimations using precision, recall, F1, and F05 for classification problems, and Spearman correlation and median absolute error for quantitative comparisons.
In addition, we utilised the aMGSIM tool to track damage at the codon level to benchmark the MMseqs2 translated searches.```
*Thread Reply:* We are still using deamSIM as NGSNGS (https://github.com/RAHenriksen/NGSNGS) still don’t have an “amplicon mode”
*Thread Reply:* but we will transition to NGSNGS as soon it is ready
*Thread Reply:* btw, I added in bioconda https://anaconda.org/bioconda/gargammel-slim in case you are just interested on the three main programs of gargammel
*Thread Reply:* This a stripped version of Gargammel that only builds the programs fragSim, deamSim and adptSim. For a full Gargammel installation look at the gargammel package
*Thread Reply:* That's awesome!
*Thread Reply:* Thanks @Antonio Fernandez-Guerra!
*Thread Reply:* this package is what aMGSIM uses
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0292077
An... Interesting one... Maybe good a #spaamtisch discussion...
*Thread Reply:* A #spaamtisch discussion on this would be great if people are up for it @Maria Lopopolo @Biancamaria Bonucci @Aleksandra Laura Pach
https://www.nature.com/articles/s41564-023-01484-x
Hi All! We have a new preprint out with @Zandra Fagernäs on dental calculus in Oceania! How does the microbiome profile compare to other regions of the world? How well do these microbes reflect migration of their human hosts? How much of the microbial communiy is as-yet unexplored? https://www.biorxiv.org/content/10.1101/2023.10.18.563027v1

Dear @channel , we propose you to fill out this form if you want to anonymously publicise a paper that you or your team are publishing or have published. The media team will look at this form once a week and post it in the community #papers channel and on twitter and mastodon if you allow it 🙂
https://forms.gle/W2vxP6cpj318RiMN6
The aMeta paper is finally out 🥳! This is a collaboration between bioinformaticians from SciLifeLab and the Centre for Palaeogenetics, involving (to name a few) @Nikolay Oskolkov, @Nora Bergfeldt, @Emrah Kırdök, me and other collaborators that are not all on the SPAAM slack! Basically, aMeta is an ancient metagenomics pipeline that takes FASTQ files as input, goes through classification with KrakenUniq, then generates a dynamic database for MALT alignment and provides several plots for authentication (edit distance, breadth, evenness of coverage, ANI, and more). It also generates an authentication score from 0 to 10 for each organism which can help you spare a lot of time so you can concentrate on the true ancient hits. Databases are available for download and the pipeline is on GitHub so feel free to try it out! https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-03083-9

*Thread Reply:* Congrats all!
*Thread Reply:* Congratulations @Nikolay Oskolkov et al.!
*Thread Reply:* Congratulations!
*Thread Reply:* Congratulation @Zoé Pochon and to all the authors
https://journals.plos.org/water/article?id=10.1371/journal.pwat.0000171
biorxiv_bioinfo@biologists.social - Revisiting the Briggs ancient DNA damage model: a fast regression method to estimate postmortem damage https://www.biorxiv.org/content/10.1101/2023.11.06.565746v1?med=mas

Congrats to @Camila Duitama and co's deCOM (featured both at a previous #spaamtisch and at this year's summerschool) has finally been published!
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-023-01670-3

Preprint from @Kadir Toykan Özdoğan
https://www.researchsquare.com/article/rs-3568244/v1
Anyone with access to https://www.sciencedirect.com/science/article/abs/pii/S1472979223000902
*Thread Reply:* Thanks @Abby Gancz however that looks really broken, the text runs off the page as does half to the images. But @Iseult already shared it above :)
Congrats to @Abby Gancz @Sterling Wright
https://www.nature.com/articles/s41564-023-01527-3
*Thread Reply:* Could I ask for someone with access to link a pdf? Thanks! and congrats! 🙂
*Thread Reply:* @irinavelsko should have one
If you are interested in how human gut bacteriophages diversified in tandem with urbanization and their associations with diseases, look at this:
"Phylogeny and disease links of a widespread and ancient gut phage lineage" https://www.biorxiv.org/content/10.1101/2023.08.29.555303v2

*Thread Reply:* Do they publish their final genomes?
*Thread Reply:* I think not yet, I will ask about it 😉
Ancient chicken viruses 👆 with @Ophélie Lebrasseur @Ophélie Lebrasseur and others
https://pubmed.ncbi.nlm.nih.gov/36744896/

*Thread Reply:* Hey @Benjamin Vernot you can update your slides saying 'HA TOLD YOU SO!'
*Thread Reply:* (admittedly this is relevant to different analysis but still
*Thread Reply:* also you might want to create super-reads before removing identical duplicates (100% identity on 100% length) so you can recover those reads that during adapter clipping and quality trimming will have shorter sequence lengths, but they are coming from the same template
*Thread Reply:* few months ago I added to aMGSIM the ability to add duplicates based on the duplicate distribution of empirical samples https://github.com/genomewalker/aMGSIM?tab=readme-ov-file#add-duplicates which uses the models from https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.13800
*Thread Reply:* hah! I love it, thanks @James Fellows Yates
https://www.researchsquare.com/article/rs-3745869/latest
Not so recent but useful for those of you interested in aDNA from South America https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10670959/
Congrats to @Piotr Rozwalak!
https://www.nature.com/articles/s41467-023-44370-0
*Thread Reply:* Fancy adding the genome to the 'Dir? 😏
*Thread Reply:* Sure 😄 , now is available in Genbank here: https://www.ncbi.nlm.nih.gov/nuccore/BK063464
Congrats to @Kerttu Majander!
https://www.nature.com/articles/s41586-023-06965-x
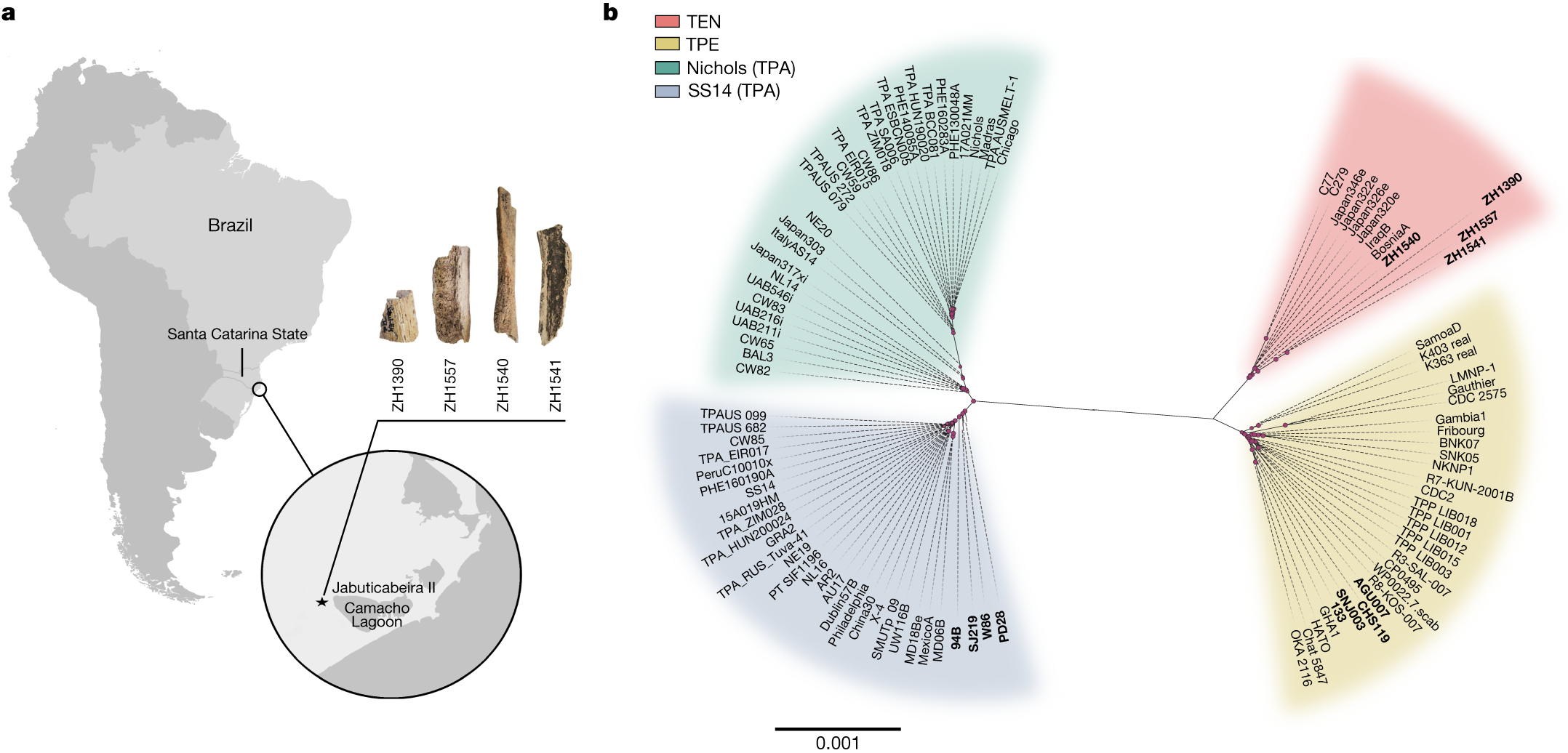
Ancient japanese palaeofaeces including@Kae Koganebuchi !
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0295924
A preprint from @Nikolay Oskolkov!
https://www.biorxiv.org/content/10.1101/2024.01.24.576930v1

Now some calculus, featuring @Claudio Ottoni!
https://www.nature.com/articles/s41598-024-52422-8
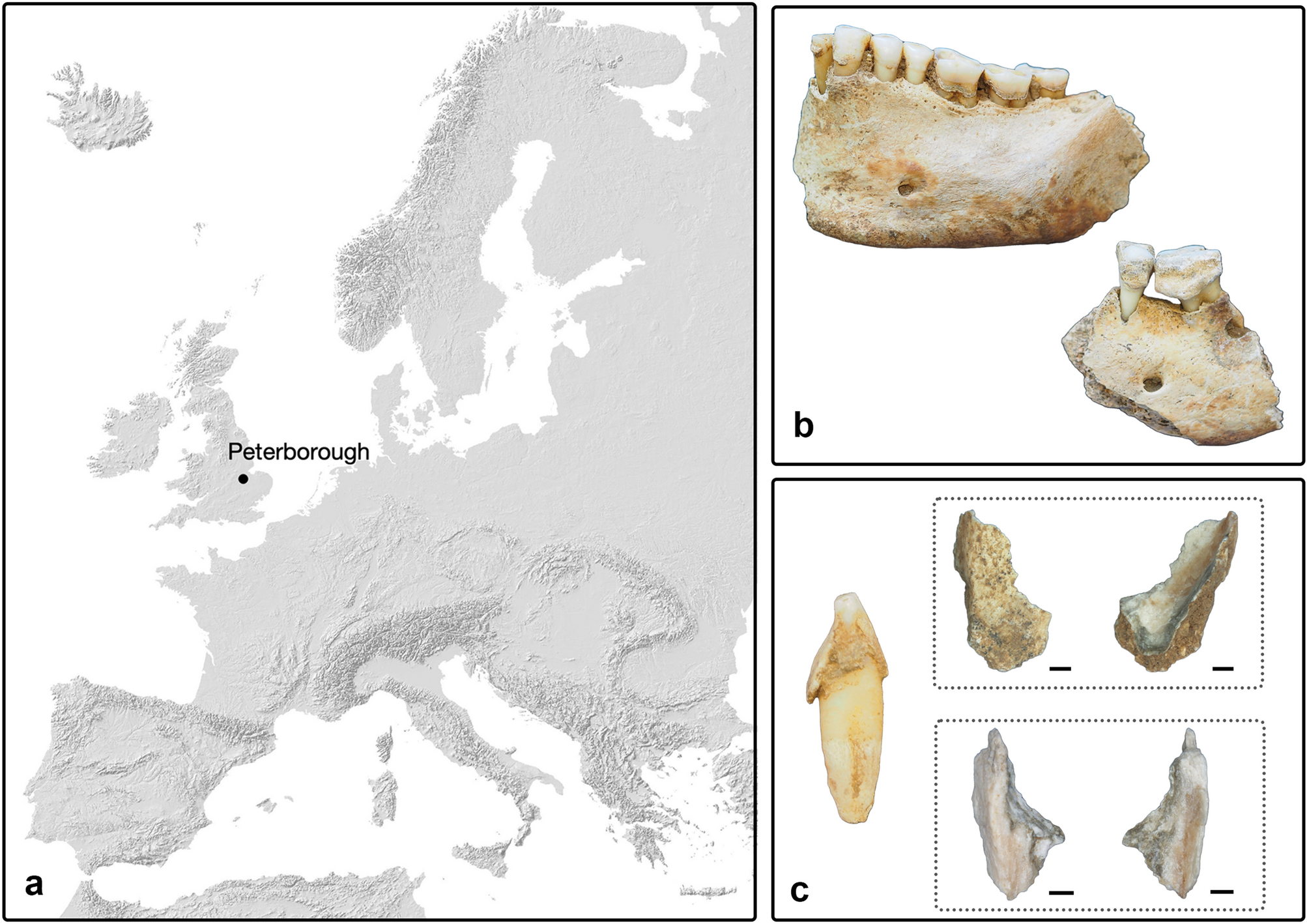
Hello everyone, I would like to share our preprint with you! Maybe interesting 🙂 https://www.researchsquare.com/article/rs-3879251/v1
Nice visualisation tool for SNPs on a MSA: https://github.com/aineniamh/snipit?tab=readme-ov-file
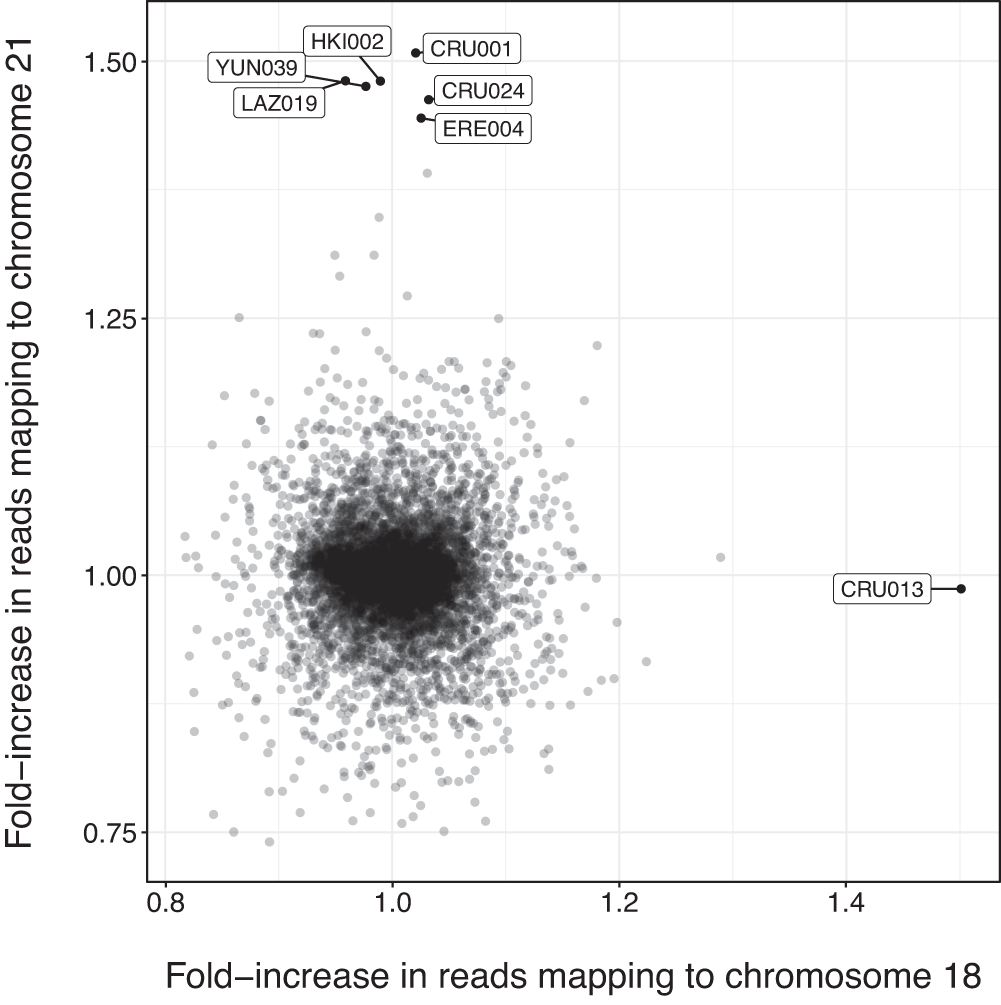
Cases of trisomy 21 and trisomy 18 among historic and prehistoric individuals discovered from ancient DNA🙀 https://www.nature.com/articles/s41467-024-45438-1
Morning all! I seem not to be able to connect to the cloud entry server, do you have this issue? @James Fellows Yates I hope you and the whole family made it through ok!
I think this is the wrong chat @Mikkel Winther Pedersen, will in the correct one
New calculus paper comparing a 16S capture technique designed for dental calculus @Raphael Eisenhofer
https://peerj.com/articles/16770/
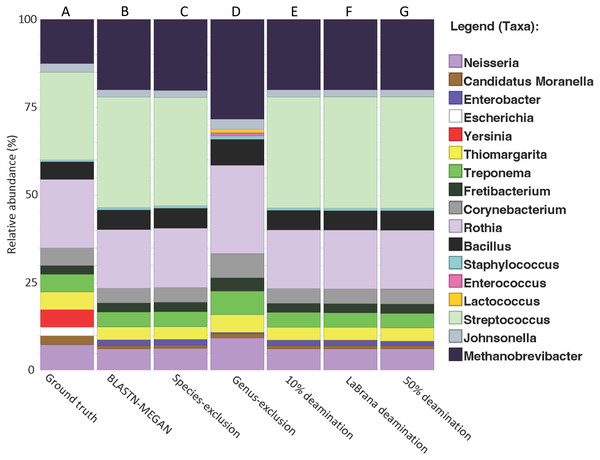
*Thread Reply:* That's a really nice study, it reads really well in terms of the order of experiments 👏👏.
@Biancamaria Bonucci @Maria Lopopolo imo this would be a nice one for a spaamtisch and a tweet, lots of Qs about amplicon/metabarcoding came up at the ACAD workshop and so this would be timely
*Thread Reply:* Thank you! And sure we can set something up. I’m hoping to defend here soon and so I’m not sure if I’ll have time until the summer but maybe Ralph would be up for this?
*Thread Reply:* Nice!! We can send you an e-mail and you guys can decide ☺️
*Thread Reply:* It would be great I agree, @Sterling Wright you could also fill this out and let us know your availability :) https://docs.google.com/forms/d/e/1FAIpQLSc0Lb3kKm2sRTutY9Nyk58SN9Lg82VfUMtVvrEmVGmwHZxw/viewform?usp=sf_link
*Thread Reply:* Filled it out!
Congrats to @Ophélie Lebrasseur @Ophélie Lebrasseur!
Ancient horse viruses!
https://academic.oup.com/mbe/article/41/3/msae017/7617356 Ancient Genomes From Bronze Age Remains Reveal Deep Diversity and Recent Adaptive Episodes for Human Oral Pathobionts by @Iseult
*Thread Reply:* Yayyyy I’ve been waiting for this! Congrats Iseult!
Congrats to @Bing Sun @aidanva and co!
https://www.nature.com/articles/s41467-024-47358-6
@Nicola Vogel is on a roll!
https://www.biorxiv.org/content/10.1101/2024.04.12.589157v1?med=mas

Recent paper analyzing biomolecular data from Roman era individuals in Romania https://www.sciencedirect.com/science/article/pii/S2352409X2400138X
*Thread Reply:* Hi @Sterling Wright congratulations!
*Thread Reply:* Since the article isn’t open-access, would you mind sharing the pdf with us?
*Thread Reply:* Thank you and for sure! I hope this helps.
Review from a Conor Meehan, who thinks the review might be of interest to us working on oral microbiomes/dental calculus
https://x.com/con_meehan/status/1790043501318537583
Paper: https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.001251

New preprint on diversity of oral Streptococcus in ancient dental calculus and modern oral samples from humans and non-human primates https://www.biorxiv.org/content/10.1101/2024.05.19.594849v1.abstract?%3Fcollection=

Congratulations @Kadir Toykan Özdoğan ! https://link.springer.com/article/10.1007/s12520-024-01999-2

A bit of self promo but here is our preprint on the eleven first ancient HHV-6A and HHV-6B genomes 🥳 https://www.biorxiv.org/content/10.1101/2024.06.25.599715v1

Plant mummies?






 }
}
 }
}


 }
}


 }
}
 }
}
 }
}
 }
}
 }
}

 }
}

 }
}

 }
}