Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
@Clio Der Sarkissian has joined the channel
@Antonio Fernandez-Guerra has joined the channel
Hello @channel nice to see the slack link is working!
@Sterling Wright has joined the channel
@channel some small updates:
1) Provisional schedule can be seen here: https://spaam-workshop.github.io/#/spaam2/programme (this will be updated over time), note the shift to afternoon/evening to accommodate the Americas 2) Session abstracts will be sent out in a week or so to help you get an idea what we will cover
@channel I’ve re-sent an email that I circulated to everyone last week since it ended up in several people’s spam folders again. Here is the contents of that email in case someone has missed it:
> Dear all, > > We are pleased to announce that the schedule, as well as session and icebreaker abstracts, for SPAAM2 are now available online (https://spaam-workshop.github.io/#/spaam2/programme, subject to minor changes!). > > SPAAM2 sessions will be held virtually on the 21st and 22nd September from 13:00 to 20:00 CEST. > > The discussion sessions will be hosted on Zoom. For those who can only listen in, this will either be hosted via Zoom (muted) or private Youtube live stream. Links and passwords will be sent on Friday 18th September via email prior to the event (please keep an eye on your junk folder). A live chat will be hosted in our slack workspace under the channel <#CPHECT30A|spaam2-open> (invite link here: https://spaam-workshop.github.io/#/?id=standards-precautions-and-advances-in-metagenomics), here all members can have parallel discussions and listener-only attendees can ask a (limited) number of questions. Please let us know if you will not be able to attend. > > In cases where it would be feasible and in accordance with the current COVID-19 guidelines for your countries/institutions, we suggest participants from the same lab-group join the meeting together on a shared call. This is to minimise the number of drop-outs in case of bad internet connections. > > We would also like to remind you that by joining any of the livestreams, you will be by default agreeing to the code of conduct, which can be found on the website (https://spaam-workshop.github.io/#/spaam2/codeofconduct). > > We would also like to circulate a list of the participants including names and affiliations to those participating in the discussions. Please reply to this email or message a member of the organizing committee on Slack to let us know if you DO NOT consent to having your name and affiliation circulated to other participants. > > Kind regards, > > Åshild Vågene, James Fellows Yates & the SPAAM2 organising committee
I wonder if someone else should send it as well, in case it address is blacklisted (ffs email sucks so much nowadays)
*Thread Reply:* I used my university email this time instead of my gmail, hopefully it should reach people now. Those who’s junk it ended up in all had uni emails
*Thread Reply:* Hopefully that helps
*Thread Reply:* Although it is ridiculous that I get more 'spam' from institutional emails then Gmail 😂
*Thread Reply:* Hopefully! If not, I hope people check the website
*Thread Reply:* Ooh we should tweet too!
*Thread Reply:* Completely forgot
*Thread Reply:* I'm on my phone with Maia at the park, maybe you could draft one?
*Thread Reply:* Yes, please do and we can retweet
*Thread Reply:* Uhh ok I can also try
*Thread Reply:* Done: https://twitter.com/jafellowsyates/status/1305528478134132736?s=19
*Thread Reply:* No problem, my mum took over
For those who are Twitter inclined: https://spaam-community.slack.com/archives/CPHECT30A/p1600097234008300?threadts=1600096599.006000&cid=CPHECT30A|https://spaam-community.slack.com/archives/CPHECT30A/p1600097234008300?threadts=1600096599.006000&cid=CPHECT30A
@channel Here come the details for the meeting next week, in case you didn’t get either of my two emails that I just sent (check your spaam folders)
```Hi everyone, (sending twice from different accounts so everyone gets it)
Below is the zoom registration link for the Standards, Precautions & Advances in Ancient Metagenomics 2 discussion workshop taking place next week on Monday 21st and Tuesday 22nd September between 1-8PM CEST. A zoom instruction manual and overview of how the meeting will function is attached (i.e. who can ask questions and how). Attached is also a list of the participants.
You are invited to a Zoom meeting. When: Sep 21, 2020 12:30 PM Copenhagen & Sep 22, 2020 12:30 PM Copenhagen
Register in advance for this meeting:
https://ucph-ku.zoom.us/meeting/register/u5Iqf-mvqTkjEtTmZHwqX9o1xht_p789LBgF
After registering, you will receive a confirmation email with the zoom link. This link will be active on both days between the times 12:30-8PM CEST.
As you enter the meeting we will add either (discussant) or (listener) to the end of your zoom name. Discussants will be able to have their video on and have full control of their microphones. Listeners will not be able to turn on their video or unmute themselves and must ask questions in either the Slack channel (#spaam2-open) or zoom chat (see attached How-to for further details).
Each session will start with one or several icebreaker talks about topics that are intended to act as primers for the following discussions. Please see the programme (note the new URL!) for a reminder of the sessions and icebreakers: https://spaam-community.github.io/#/spaam2/programme
We would also like your input! Please let us know ASAP via slack or email if there are any topics that you would like to be discussed. We will not have a set agenda for the discussions, but will have a slide(s) with a list of relevant topics/issues for each session that we can draw upon if people are feeling shy ;). We would like to add any suggestions you have to these slides. Please indicate a relevant session your question/topic could be talked about. We will also try to avoid discussions of one topic lasting for too long.
Additionally, in the last few days we have had a lot of last-minute requests from people who want to listen in/watch. However, since this is not a normal conference, but a discussion-based workshop where we encourage participation from people (particularly PhDs and postdocs) who are actively doing the research, we, the organizers, have decided not to allow further participants as we are already 77 people (38 discussants and ~39 listeners).
If you know someone who would like to listen-in we will allow them to share your stream (if the current covid-19 guidelines in your area allows this).
We will be removing meeting registrants whose names do not match up with the participant list attached.
See you on Monday!
The Organization Team Standards, Precautions & Advances in Ancient Metagenomics 2```
@James Fellows Yates Is the sky blue in Jena? (test)
@channel - please feel free to change the ‘View’ setting on your zoom - on the top right corner. You can change is so you can see the presentation speaker alongside the presentation etc
@channel any questions or comments for Zandra or Jaelle specifically?
I am working with soil samples and there isn't much that we can do to minimise the contamination. Any thoughts about that?
*Thread Reply:* Will get to that after Irina's initial discussions
*Thread Reply:* Probably discussion points 2
Depending on your sequencing centre, you might not be able to track down index hopping, if the indices have been removed as part of pre-processing by the centre
It works well if you have in-line barcodes. Then quantification of index hopping is easy
Sooooooooooooooooooooooooo you guys were too enthusatic in voting and so I'm going to change polling app.
Should we sequence all the blanks? Or only 1 per batch (we are usually having 2)?
OK the poll also reads markdown 🤦 The second '2' is meant to be \>2
What about library prep blanks? Sequence those? How many per lib prep batch?
1 extraction + 1 lib blank per 10 samples usually
*Thread Reply:* that's what I used to do as well
We work with historical samples: not old enough to have a lot of damage. We are really happy about any damage we find to use for authentication
We tend to sequence deeper as far as I know (I did this), and we've been playing with bioinformatic ways of dealing with damage
Wouldn't sequencing blanks in a separate run create additional batch effects?
*Thread Reply:* You'd think ;). But the reason why we ended up doing that because we had leaking of capture libraries into blanks
*Thread Reply:* And people got very Paranoid
*Thread Reply:* Ah, makes sense. With in-line barcodes that's not a problem. They have other problems, though, like reduced ligation efficiency depending on the barcode sequence
I hadn't heard of internal barcodes until now...
*Thread Reply:* I have, but still feel very fuzy about them!
Please note that there are ligation biases with internal barcodes. Check out Brealey et al. 2020 MBE
*Thread Reply:* Awesome thanks! Could you please link the paper?
*Thread Reply:* I think the barcode bias discussion is in the supp
*Thread Reply:* Yes, we had to move it out of the main text so it's completely in the Supp Info document
Related to the genotyping and the issue of using just a single reference genome, there is this paper from last week that propose a nice turnarround by mapping into graphs rather than genomes... https://pubmed.ncbi.nlm.nih.gov/32943086/

Using this in the framework of ancient microbes could be quite interesting
*Thread Reply:* I was going to ask a question about this very paper 🙂
I think this is the Hungarian mummy paper: https://www.nature.com/articles/ncomms7717

@Kun Huang do you know how much modern data there is for M. oralis?
*Thread Reply:* There's four modern genomes available
*Thread Reply:* As I know, there are only 4 modern RefSeqs of M. oralis, and maybe two modern calculus samples which contain well-covered M. oralis. These modern samples are all from @velsko or Mann et al. study.
*Thread Reply:* Thinking in terms of the variation graphs what would be good test data. Pathogen stuff obviously pretty good for that, but outside of that comes more difficult
Has anyone else used the GreenGenes reference or RefSeq for their MapDamage analysis? I applied GreenGenes once before with dental calculus samples and was able to get damage plots.
*Thread Reply:* But I wasn't very confident in the results.
*Thread Reply:* Hi Sterling...why not confident?
*Thread Reply:* There were about 10,000 reads that mapped but because those references are mixed with soil, oral, skin and all sorts of microbes, I wasn't sure how to interpret it. If I were to go back, I think it may make more sense to use the HOMD database as a reference for MapDamage. Has anyone tried this yet with dental calculus?
*Thread Reply:* @Lena G @Pooja Swali what do you use?
*Thread Reply:* htsbox majority consensus caller by Heng Li so far, but haven't done a careful evaluation
*Thread Reply:* Oooh, not heard of that. What is the possible advantagE?
*Thread Reply:* Angsd, but I've only used it for single genes so far.
I've also pinned the discussion points to the the channel! To see this, please press the 'i' button in the top right hand side of the chat (next to the display pictures)
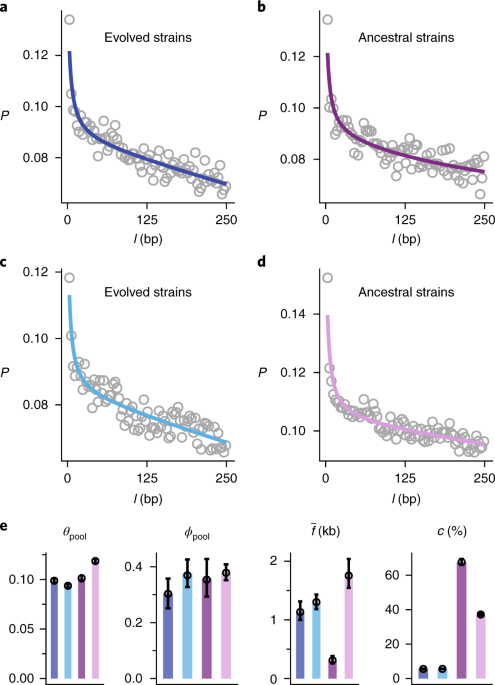
I think it’s worth considering the definition we use of strain. In the standard sense it suggests members of the same species that have different gene content. Here, it seems to mostly mean members of the same species with (enough) SNP differences. How does this affect the way we think about differentiating strains and the methods we use to do so?
here is the very fresh & through review about strain lineation etc… https://pubmed.ncbi.nlm.nih.gov/32499497/

What is the benefit to using freebayse over MultiVCF Analyzer?
In case of use, this is a nice tool for reconstructing pan-genomes from a modern repertoire: https://www.biorxiv.org/content/10.1101/2020.09.11.293472v3.full
*Thread Reply:* I have a bit of experience with panX and also roary. Both easy to use but requires tweaking to incorporate ancient data
*Thread Reply:* PanACoTA is an excellent name
Anyone using DeepVariant ? https://github.com/google/deepvariant
*Thread Reply:* ```DeepVariant supports:
Germline variant-calling in diploid organisms. For somatic data or any other samples where the genotypes go beyond two copies of DNA, DeepVariant will not work out of the box because the only genotypes supported are hom-alt, het, and hom-ref. The models included with DeepVariant are only trained on human data. For other organisms, see the blog post on non-human variant-calling for some possible pitfalls and how to handle them.``` sigh
A nifty comparison of SNP calling pipelines for bacteria
Here are potential alternatives to MultiVCFAnalyzer that I’ve been playing with: https://github.com/Integrative-Transcriptomics/MUSIAL https://github.com/tseemann/snippy https://github.com/sanger-pathogens/snp-sites
*Thread Reply:* snp-sites works pretty well and extremely fast.
*Thread Reply:* And this one too https://github.com/Amine-Namouchi/snpToolkit
*Thread Reply:* how have you found snippy with aDNA?
*Thread Reply:* Haha I have only used snippy for making multi-sample SNP alignments because you must use their processing pipeline with little power to modify parameters. So I input bams that have been prepped using aDNA standards.
*Thread Reply:* yeah it is designed as a full pipeline so I was wondering how it does with aDNA bams and core snp alignments, but I guess we are back to lacking configuration options...
*Thread Reply:* A few of us in the poinar lab are using it like Kelly was saying
@channel has anyone tried more recent aligners? Like Minimap2/
It's designed for but I'm pretty sure Heng said it has similar perofrmacne
Has anyone had any experiences with aDNA specific genotypers: e.g. antCaller?
https://github.com/BoyanZhou/AntCaller
Last call for questions for the entire session @channel!
Also for people just listening in!
Here is a tool to detect recombinant region: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004041
detecting recombination from metagenome data : https://www.nature.com/articles/s41592-018-0293-7
ClonalFrameML works well when genome-alignment is small (30-50 taxa). The second one is pretty fast even for large-scale metagenomic datasets.
@channel does anyone of the workshop participants know MALT developers? Would like to have contacts to ask a few questions
*Thread Reply:* http://megan.informatik.uni-tuebingen.de/
*Thread Reply:* There is a dedicated section for MALT
*Thread Reply:* Ok, thanks @James Fellows Yates I think I tried that one, did not get answers on my questions, just wanted to see if somebody in the workshop is actually a MALT developer
*Thread Reply:* Ah no, they aren't present here.
*Thread Reply:* But it's basically just Daniel who develops afaik
*Thread Reply:* Ok, thanks! Maybe I can throw my question to the community when touch MALT at this workshop?
gubbins is also widely used on bactieral data
*Thread Reply:* I would just like to comment on how wonderful the name gubbins is.
But gubbins can only detect the recombinant stretch between taxa given. ClonalFrameML nonetheless can detect any recombination introduced. Just a bit computational-demanding with data growing.
I tend to use cfml, what I have observed with gubbins is that it has a tendency to output much smaller intervals, where cfml will cluster them and take into account non-core intervals
@channel starting in 2 minutes!
I'm using Refsoil as a database for soil contam (I don't have a soil sample from this site 😞 ) and was curious what others are using database-wise
I would also like to know what people with “unconventional” data sets (ie not human calculus) that do not have good reference databases to compare against in Source Tracker or your favourite tool?
we have developed a workflow that exploits the MGnify API to generate training sets for mt-ST. It uses the MGnify biome information to query data from MGnify. Now we are implementing the training of mt-ST for ancient samples
check slide 3 from here, we are going to make it public soon
My samples come from extreme environments and specially for thermophilic or acidophilic species I don't see any DNA damage. Of course I wouldn't do any big assumptions right now because I am still working on that. Also there a lack of reference genomes for this type of species
but is definitely something to look into
We have also some palaeofaeces samples from very arid environments and see almost no damage either despite the samples being dated to 1300 BP.
So water is necessary to accumulate aDNA damage patterns.
It makes totally sense. Definitely the lack of aDNA degradation patterns in microbial data is highly frequent. It would be something to quantify somehow. I think for the field is something that will help many, to have an article to reference to when degradation patterns are not seen in our datasets.
we are exploring ways to circumvent this problem by exploiting the differences of codon usage of the sequences in our samples
*Thread Reply:* You mean regarding the lack of aDNA patterns?
*Thread Reply:* it might not be a universal solution but for certain situations might be useful. The stuff we are doing is inspired by this https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3799439/

*Thread Reply:* The only problem I could see is how to persuade the reviewer that is a non-standard, but valid approach. 😉
*Thread Reply:* microbes and their special needs 🙂
*Thread Reply:* there is a lot of info hidden in the coding space that we can exploit in combination with the methods of @Nikolay Oskolkov
*Thread Reply:* Definitely, there is a lot of reference data to train for codon usage.
*Thread Reply:* when we don’t have references, we use a super-read like approach to partition potential ancient reads
there are certain signatures that might be useful when there is no damage
We also use this as an alternative to Mapdamage but is mainly useful for samples that are UDG treated https://github.com/pontussk/PMDtools
@James Fellows Yates James, are you suggesting that we need something like the standard PCA plot standard for the pop gen folks?
*Thread Reply:* Will ask next!
We all look terrified now.
I would say you can compare but need to record the metadata, and do some statistical modelling taking them into account. Combat and Songbird are nice tool for that https://github.com/biocore/songbird https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2263-6
I agree, it is time to introduce generalized linear models into microbiome analysis.
yeap that;s something that is going to be very useful!
https://evayiwenwang.github.io/Managing_batch_effects/
@Maxime Borry and @Alex Hübner all batch-effects correction tools will work providing that the signal is orthogonal to the batch. With 100% confounding when reference and query samples were sequenced using different platforms I would not put them on the same PCA plot :)
*Thread Reply:* Agree, can’t compare 16s and WGS data for example, or metagenomic profiles generated by classifying agains different databases, or different tools. As an optimistic note: keeping track of the metadata and visualising their effect, on let’s say a PCA colored by batch effect, will give a good starting indication of whether or not this is something to worry about
*Thread Reply:* True! I usually make a correlation heatmap of my PCs against all available meta-data to understand the sources of variation in my data. If my PC1 is mostly explained by the batch variable, it is very worrying
Last one on batch effect correction for tonight: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1850-9

@channel I've just made a channel for people working on ancient microbial genomics (e.g. single genomes of pathogens, commensals etc). <#C01B511KU91|microbial-genomics> . Please join if you work in this area!
Code of conduct: https://spaam-community.github.io/#/spaam2/codeofconduct
Even though a side effect of unethical practices is data which is not publicly available, I think it should be considered that many Indigenous groups do not want their data to be public.
*Thread Reply:* But is it in this case ethical to work on that at all?
*Thread Reply:* I think it depends on the group.
*Thread Reply:* What is the motivation behind making the data not public?
Both the side effects and by extension the solutions are not one size fits all in this case BUT I think as Miriam said it will take increased interaction and involvement with existing descendent communities for Indigenous communities to practice ethically.
the data will depend if it is under the Fort Lauderdale Agreement, in addition if the country is under Nagoya
Fort Lauderdale Agreement: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2672783/

of course this is for genomic data, no idea about other types of data
one gets lost with all this stuff
Yes but I would argue that these and also NAGPRA fall short so we have to take a bit more responsibility in this.
there is also a time limit - ie when samples were collected , if they are in museums abroad for decades before being sampled etc
*Thread Reply:* Especially museum samples collected along time ago, e.g. during colonial times are a delicate issue.
I guess there are the needs of aligning all sites involved
Questions for voting for Nico - Put emoji reactons to those you are most interested in!
"Data clean-up: How far can we go?"
I guess having a DIR of institutions (legal, ethical…) would be also very useful
"Taxonomic levels: What’s the right size for the lens?"
And I copy here the other slides in case it helps refreshing the sub-items of each point in the poll
A note on radiocarbon reporting conventions
Regarding SOPs, these are some items we wrote with James: Benchmarking: What analyses should everyone do? (e.g., compare to source-tracker) What validation tests should be run? What data should be used? Which datasets for training (e.g., for machine learning)
maybe we should have our own branch of CAMI: https://data.cami-challenge.org/ maybe we should contact them
*Thread Reply:* @Maxime Borry has spoken with the person leading this but found she wasn’t very interested. Maybe having a group collectiveley approach would be more persuasive
*Thread Reply:* Yes, I can get in touch through the people from my former group in Bremen, when CAMI appeared they contacted us
*Thread Reply:* at that time was Alexander Sczyrba
*Thread Reply:* Well, let’s say that no one went forward with it.
*Thread Reply:* There is also the LEMMI benchmarking challenge that is quite interesting, because it is updated in real time https://lemmi.ezlab.org/#/ We contacted them, but it also didn’t go forward https://gitlab.com/ezlab/lemmi/-/issues/5
*Thread Reply:* This should be coupled with the lab standards @Christian Carøe was talking about. For example the guys from the XMP develop this https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5345951/ In last year GSC meeting Scott presented their standards

This is the paper Jamesis referring to: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003531
*Thread Reply:* great paper! I had the same issue James (i.e. how to norm? etc.)
And this one: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02224/full
Speaking as a PI here: Testing is super important, but I'd want my people to come out of my lab with a portfolio that would allow them to pursue whatever career they want. If it is a career in science, they will need publications. Testing will not result in a pub...
*Thread Reply:* But then who should do the testing? I would disagree testing doesn't result in a publication..., 50% of bioinformatics is just hat 😆
*Thread Reply:* Bioinformaticians with permanent positions, e.g. as those offered by SciLifeLab
*Thread Reply:* Permanent positions?! They are a thing!?!?! 😉
*Thread Reply:* Positions in core-facilities are not necessarily permanent 😉
*Thread Reply:* Those at SciLifeLab are 🙂
*Thread Reply:* I was also referring to what I myself have an interest in - it's not so much whether I will get a publication out of it, it's also what I'm interesting in doing, which is more data analysis, not testing. But I agree testing is super important and useful!
*Thread Reply:* I think that testing is good and necessary, but it is necessary to put an end at some point, choose one solution we believe that will satisfy reviewers and community, and focus on the questions of the project. I personally care much more about questions than methods. So as long as the method is sounding, although not perfect, I prefer to move on
*Thread Reply:* I also find it interesting (@Katerina Guschanski), although you say testing will not result in pub, there is clearly a need for these testing papers as it seems one of the biggest concerns of people (particularly from the session yesterday). So if someone was to do that, and publish it, in principle they could get lots of citations?
*Thread Reply:* Absolutely true and a super good point. I/we would be happy to be able to use such guidelines. Also perfect arguments to provide to reviewers when they go "Why didn't you use this or that tool...?". So I guess the truth is somewhere in the middle. Periodic testing of new tools to compare to winners from the previous round?
*Thread Reply:* Exactly, if we had a CAMI-like dataset, so a 'gold standard' for people to test - would you feel more comfrotable for a student to spend time on it?
*Thread Reply:* In the sense there is already a precedence
*Thread Reply:* for this we need a way to generate ancient mock communities in the lab, no idea if this is a thing
*Thread Reply:* Nope, I don't think we could do that; unless we get a single Mammoth femur and freeze it
*Thread Reply:* But we would need stuff for different contexts; this isn't just microbes
*Thread Reply:* one step at a time
*Thread Reply:* That would be perfect (using a mock community for tool testing periodically). Ideally coordinated effort across labs with multiple participating researchers.
*Thread Reply:* Wouldn't it be possible to produce a "simulated" community with typical aDNA traits: Damage, contamination, fragmentation patterns, etc. It is possible for hosts...
*Thread Reply:* Yeah I guess. IIRC CAMI had both synthetic dataset and also a 'real' mock community (with spiked in stuff)
*Thread Reply:* yep the real stuff is the interesting
*Thread Reply:* Exactly, we can't really model aDNA datasets very well atm
*Thread Reply:* the synthetic should not be a problem
*Thread Reply:* Well, we don't know the ground truth of a real aDNA dataset at the moment 🙂
*Thread Reply:* Yeah like Gargammel uses a Neanderthal damage profile... because everyone is analysing that...
*Thread Reply:* you, can use your sample patterns, but I think we need more realistic models
*Thread Reply:* @Nikolay Oskolkov I guess we could use some DL to generate more reliable syntcomms
*Thread Reply:* You mean even more complicated than what we currently think is already nightmare 🙂?
I disagree. Publications are only necessary if one goes into academia
This kind of testing would be great for someone who wants to go into industry, especially in bioinformatics work which is in high demand
Agree with @ivelsko. That's why I said career in science (meaning academia, sorry, should have been more precise).
For those in Germany you have https://www.gfbio.org/ for this
and the standard: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4511511/

Notorious C.P.H
Yes for some funding bodies and journals requiring data upload
*Thread Reply:* > Examples of activities that alone (without other contributions) do not qualify a contributor for authorship are acquisition of funding; general supervision of a research group or general administrative support; and writing assistance, technical editing, language editing, and proofreading. Those whose contributions do not justify authorship may be acknowledged individually or together as a group under a single heading (e.g. “Clinical Investigators” or “Participating Investigators”), and their contributions should be specified (e.g., “served as scientific advisors,” “critically reviewed the study proposal,” “collected data,” “provided and cared for study patients”, “participated in writing or technical editing of the manuscript”).
As far as I know, there are no structures in place to enforce or penalise authors/researchers who do not make data publicly available. Neither from journals nor from funders. These are general requirements, but even if journals are alerted to the violation of this rule, they cannot do anything about it (or at least don't do anything).
Also the CREDIT one: https://casrai.org/credit/
from peerJ https://peerj.com/about/policies-and-procedures/#authorship-criteria
*Thread Reply:* > Acquisition of funding, collection of data, or general supervision of the research group alone does not constitute authorship.
Most modern microbiome samples do not include human DNA
*Thread Reply:* How did you estimate this? I disagree from my experience with them
*Thread Reply:* I think SRA may screen and mask the human reads
*Thread Reply:* Stripped before uploading to ENA
*Thread Reply:* \don't know 😆
*Thread Reply:* Or you have to apply to some ethical body that sometimes they come back to you and sometimes not. E.g. dbGAP.
Most common question I used to get from curators : “Why should i give you the sample and not wait for the next technological revolution in 10,15,20 years?”
*Thread Reply:* Second that!
*Thread Reply:* Because we can 'immortalise' libraries 😉 (according to the mantra of a certain JK)
*Thread Reply:* Hahaha! I admit to have used this argument in the early years...
*Thread Reply:* but if the data cannot be compared to anything else (from our conversation yesterday) is it really a bioarchiving of samples?
*Thread Reply:* It did come from the same source
*Thread Reply:* This is really interesting - thanks for asking!
I think it's also important to remember that many archaeologists are working from an export permit where they specified research agendas that may not be in line with all the results metagenomic approaches can lead to.
For example working with pathogens.... if our archaeologist specified TB in a research proposal and export permit but we found something else, we cannot reconstruct that and work with that.
In some cases
I've shared it with the other organisres, someone else will clear it up 😉
@channel if there is anyone not comfortable sharing their picture in the group photo on twitter, please let one of the organisers know
If anyone missed the group photo, please let me know
And we will take single-ones later and do some photoshop magic
Assembly+binning is a standard analysis in modern metagenomics. We are hesitant to do it in ancient metagenomics because assembly is a very complex step that needs lots of reads, do we even have enough coverage in ancient microbiome?
*Thread Reply:* Depends on the sample; bigger problem is short read lengths
*Thread Reply:* not always, there are other alternatives than MAGs
*Thread Reply:* It worked well for us for some (highly abundant) species from historical dental calculus samples
*Thread Reply:* i work with very complicated samples, and is very challenging
*Thread Reply:* so the traditional approach doesn’t work
*Thread Reply:* this is like going back 10 years back in time in metagenomics
*Thread Reply:* How do you handle the aDNA damages during the the assembly?
*Thread Reply:* we are working on integrating this in the assembly process
*Thread Reply:* I will show briefly
*Thread Reply:* also not using de brujin approaches
there are some papers where they did de novo assembly of microbial genomes
*Thread Reply:* very true but it was targeted - not an open exploration. There are only a handful of papers fidning ancient MAGs
*Thread Reply:* One of the few recovering ancient MAGs by @Katerina Guschanski and @Jaelle Brealey https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msaa135/5848415
*Thread Reply:* The limitation is: these are very recent samples my most standards (within the last century) and the used approach worked for highly abundant species
*Thread Reply:* Exactly! Hence the need for the combination of assembly plus assembly free methods
*Thread Reply:* but it opens very exciting possibilities
The one on dental calculus from C. Warinner is an example
Ok, I understand that doing assembly+binning you will discover new unknown microbes. How would you interpret and analyze those microbes?
*Thread Reply:* for example http://merenlab.org/2016/11/08/pangenomics-v2/
*Thread Reply:* we have a full workshop every year on this
*Thread Reply:* https://pagesperso.univ-brest.fr/~maignien/ebame5.html
*Thread Reply:* For Maxime and me, we mainly work on very well described datasets, like human gut or human oral microbiome, which makes it easier to compare to known data. For unknown environments, it is more complicated.
*Thread Reply:* @Antonio Fernandez-Guerra Is it too late to sign up for the workshop for this year?
*Thread Reply:* I would also be interested in attending this workshop
*Thread Reply:* it is an excellent workshop! But sadly i dont think it is happening this year.
*Thread Reply:* however keep your eyes on twitter or email Louis for further info for when they host it next: lois.maignien@univ-brest.fr
*Thread Reply:* Also bear in mind this is not geared for ancient metagenomes
*Thread Reply:* Thank you @Anna F.!
*Thread Reply:* there will be some online courses available early next year if that would be interesting? again not necessarily ancient
*Thread Reply:* I would still be itnerested. Are they hosted by the same people?
*Thread Reply:* partially! Tom Delmont will be on the teachers. It will be a short course so an intro to the field
*Thread Reply:* Next year unfortunately
*Thread Reply:* @Nikolay Oskolkov this one https://www.biorxiv.org/content/10.1101/490078v2 from Simon Rasmussen is using Variational Autoencoders
*Thread Reply:* CONCOCT from Chris Quince https://pubmed.ncbi.nlm.nih.gov/25218180/ works very nicely

*Thread Reply:* this last one from Chris is also super cool https://www.biorxiv.org/content/10.1101/2020.09.06.284828v1.full

@Maria Spyrou https://pubmed.ncbi.nlm.nih.gov/31235882/ PLASS

PenguiN: https://twitter.com/thesteinegger/status/1306245987048910849
now we are trying to model damage during the assembly process
we have ways to cluster co-abundant genes that might come from the same population
@Nikolay Oskolkov what kind of artefacts did you get for those early assemblies? How did they not work?
@Nico Rascovan @Antonio Fernandez-Guerra what would you like your channel to be called?
*Thread Reply:* What do you mean? Any channel, or something related to what we discussed today with the assembly of aDNA data? Antonio knows about it, I am just interested on it, but I'm still very ignorant on this respect
*Thread Reply:* I would add one like New computational methods or something similar, where we can discuss new approaches
*Thread Reply:* analysis-hipsters? ;)
*Thread Reply:* also one channel with something like Matchmaking for potential new collaborations
*Thread Reply:* You guys should be able to make these channels actually 🤔. Make them and advertise on #general! If you can't let me know
about unknowns… (shameless self promotion) http://merenlab.org/2020/07/01/dark-side/
The papers that describe the assembly methods used by the Segata lab are: https://www.sciencedirect.com/science/article/pii/S0092867419300017?via%3Dihub
https://www.sciencedirect.com/science/article/pii/S1931312819304275?via%3Dihub
With P. copri has ancient genomes
*Thread Reply:* Yes, but very fragmented. ~40% region is confidently covered.
http://merenlab.org/2016/12/14/coverage-variation/
Has anyone compared assemblies of UDG-treated and non-treated samples?
but it will be interesting to actually see the comparison
@Maxime Borry showed that the damage doesn’t make a difference, only the read length does
But we didn’t simulate UDG-treated data, only non-treated.
@channel please post in the thread to this message any papers you know they performed assemblies of aDNA data!
*Thread Reply:* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5643016/ Luhmann 2017

*Thread Reply:* Segata lab poop: https://www.sciencedirect.com/science/article/pii/S1931312819304275?via%3Dihub
*Thread Reply:* Jaelle reindeer/bear/gorilla calculus: https://academic.oup.com/mbe/advance-article-abstract/doi/10.1093/molbev/msaa135/5848415
*Thread Reply:* hep B: https://elifesciences.org/articles/36666
*Thread Reply:* https://www.nature.com/articles/ncomms3172
And we can make a bioliography
*Thread Reply:* Segata lab poop: https://www.sciencedirect.com/science/article/pii/S1931312819304275?via%3Dihub
*Thread Reply:* Jaelle reindeer/bear/gorilla calculus: https://academic.oup.com/mbe/advance-article-abstract/doi/10.1093/molbev/msaa135/5848415
*Thread Reply:* assembly for y. pestis: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5643016/

*Thread Reply:* Assembly for ancient S. enterica Paratyphi C https://www.cell.com/current-biology/pdfExtended/S0960-9822(18)30694-8
*Thread Reply:* A couple from the lake sedaDNA world: https://www.biorxiv.org/content/10.1101/2020.04.10.035535v1 https://www.biorxiv.org/content/10.1101/2020.01.06.896068v1


As I know, there are very few. I know a very bad one
*Thread Reply:* 😅 sorry I panicked as people were posting on different ones
*Thread Reply:* now there are several threads...
*Thread Reply:* 🤦 James fail
*Thread Reply:* I'll collcet them all in one place 😅
Hey all, I am 100% positive about any intiative to continue and develop further the SPAAM community. I won't be able to attend at 19h, because I am taking care of my daughters now. It was a great e-meeting and I am very happy to have seen you all. See you all soon, I hope!
*Thread Reply:* Feel free to post (if you have time) specifci things you would like to have/see
*Thread Reply:* Hey, same for me here, I wont be able to make it to the last session. Thanks so much for organising and it was nice meeting you, also very interested to continue the communication in the future - I really like the idea of little workshops, maybe on assemblies :)
Just as a note, I'm new to assembly but, I tried denovo for a single genome with min30 vs min45 bp using SPAdes and found that min45 performed slightly better (assessed with QUAST and BLASTing post assembly). Min35 had more 'non-target' DNA (though I didn't filter out human, nice point on that today).
*Thread Reply:* Good to know! i’ve seen both to be honest - filtering out the host and not filtering out the host- it really seems to depend whose modern metagenomic papers you come across!
@channel remember everyone can unmute themselves and contribute in the final session 😄
@channel we are about to start, webcams on please! Listeners as well!
Here is a link to a 'metagenomics reporting guidelines' paper just out, so there is precedent for this type of paper for modern datasets. Something similar could work for aDNA and I would think would be widely cited https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30199-7/fulltext
The workshop on reproducibility that we hosted at the Max Planck in Jena last year: https://rrdm-shh.github.io
Just another example I mentioned https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000335
Got to go, but thanks very much everyone! Looking forward to digesting all of this.
Likewise-- thank you all! Excited for everything to come!
Thank you so much everyone!! This was GREAT! 😁
Really enjoyed the workshop, the organizers have done an outstanding job, thanks a lot everyone!
Thank you everyone!!
Thanks all so much for this, especially the organisers for putting this together! Hope to see everyone in person soon!
Thank you to everyone for interesting talks and discussion and especially the organisers of course! Very reassuring to hear others often feel alone - but look at us all! 💪
Thank you everyone! This was a terrific experience. Big congratulations to the organizers who pulled this off! All y’all are improving the field as a whole
Thank you everyone for this great meeting!
Thanks for hosting a great event, apologies I couldn't attend for more. I got a lot out of it and am excited to see what comes out of the community and discussions 😁
@channel we will keep this channel open for a few more months until notes are shared etc, but in the meantime Irina and I have created the different channels for all the working groups that people said they would be interested in talking in (and add those who put their names on the google sheet):
Working group on standardising metadata <#C01BX7EM4EL|metadata-standards> CAMI challenge-like benchmarking datasets/challenge <#C01BDHQP3EY|benchmark-datasets> Standardising authentication criteria <#C01B7J8M2JX|authentication-standards> Open letter on data sharing #datasharing Workshop on data reproducibility (conda, docker, github etc. etc.) <#C01B7CA42AE|analysis-reproducibility> Lab comparison: Broad lab extraction/library building testing w/ the same sample <#C01BJPKHH9P|lab-community> Cross-lab pipeline testing (e.g. genome reconstruction, or taxonomic profiles) <#C01B49RM353|analysis-comparison-challenge> SPAAM3 <#C01BL1HD57T|spaam3-organisers>
Remember these are open, you can join/leave when you want. And it is up to the people in each channel to decide how to run things! We look forward to seeing what comes out of these 🙂.
And finally, some general discussion channels:
Ancient microbiomes <#C01BDJBM1PE|ancient-microbiomes> Ancient microbial genomics (e.g. pathogens) <#C01B0LND80N|ancient-microbial-genomics> seda/environmental DNA <#C01B7CRJK7U|seda-dna>
and to add: De novo assembly and genome resolved metagenomics (e.g the unknown fraction of ancient metagenomes and what was covered in session 5) <#C01B7FPCBKL|de-novo-assembly>
Hi guys, do not know what channel to address this question to, so posting it here. Does anyone have a way to work with the SAM-alignments produced by MALT aligner? There is a known issue with CIGAR string for a number of alignments that MALT generates, that makes the SAM-file produced by MALT impossible to be processed using samtools. I can use bash/awk or python for manual processing the MALT alignments but wonder maybe there is an easier solution? Maybe @Alex Hübner or @James Fellows Yates can help? Thanks!
*Thread Reply:* I’m having the same issue with DIAMOND-produced sam files. I’d also like to know if anyone has a solution
*Thread Reply:* Hace you used the sparseSAM output? If so I would recommend turning that off
*Thread Reply:* If you didn't use that I need to check if I did anything special in a different project I was using them recently
*Thread Reply:* I don't think I did though
*Thread Reply:* No, the issue w/the DIAMOND sam files is that it’s a pseudo-sam file, since they aren’t designed for protein alignments. @Nikolay Oskolkov did you run MALT in BLASTX mode? Do you also have protein alignments?
*Thread Reply:* @ivelsko I ran it in BLASTN mode for aligning DNA reads. @James Fellows Yates I did not use sparseSAM, no, here is how I ran it:
malt-run -at SemiGlobal -ssc -m BlastN -i fastqfile -o fastqfile.rma6 -a fastqfile.sam -t 20 -d maltdatabase
*Thread Reply:* Hmm, ok that looks pretty similar to what I use but I think without the soft flipping (iirc that's what the SSC flag does?). I'm having a day off today, but if you ping me tomorrow I'll check to see what I did
*Thread Reply:* Was there any particular error samtools was reporting?
*Thread Reply:* Thanks @James Fellows Yates, you are right, the issue seems to be related to the soft-clipping. I will check without the -ssc flag. Still strange that if you want to implement soft-clipping, you get a strange sam-file that can not be read by samtools
*Thread Reply:* Yeah, there are a few wierd things about the malt implementation. I would still try posting on the megan forum again. Semester is starting up again so maybe the Devs are back at work. It took me one or two attempts to get replies previously
*Thread Reply:* yeah I don't see myself doing anything special in my code
*Thread Reply:* But I don't think I had to touch the cigar string
@channel Find here the draft of all the links/references posted during SPAAM2: https://docs.google.com/document/d/1oXS0MHg1DdB0xe3vlen5TEcC2SFZsp1aSwEOnK_soBM/edit?usp=sharing
Consider this a semi-annotated bibliography (I tried to put the context of each link, as far as I could remember/reconstruct from the chat), which we will bundle with the notes/slides from SPAAM2 and will make public.
Please feel free to make edits etc!
@channel I'm just preparing the participant list for the notes/slides that we will be making publicly available (coming for your comments/corrections on Monday!)
We've used the Zoom records to take people who were present. *Is there anybody who shared someone else's connection and therefore didn't log in themselves?*
Yes, @Markella Moraitou
Hi @channel, Clio sent a follow-up email to the SPAAM2 workshop yesterday, be sure to check your spaam/junk folders for the email
*Thread Reply:* Real time Rapid markdown fixes, love it 😂
Below is the contents of the email: ```Dear all participants,
Thanks to you all again for taking part in the online meeting.
We are happy to share with you some of the meeting outputs:
- the list of participants,
- the abstracts, notes and poll results for each session,
- the sessions' slides, including those of the icebreaker talks that the speakers were happy to share,
- the list of references and resources circulated during discussions. They can all be found in this Google Drive folder: https://drive.google.com/drive/folders/1XPITeI2Sku9WScE85d4suyYXK1OyFgpU?usp=sharing
If you wish to make any comment, change, correction, suggestion, addition to the notes and references, we encourage you to do so directly on the commentable files in this shared folder before Friday October 16. After this date, the files will be locked and uploaded onto Zenodo as pdf for long term archiving (and include a citable DOI).
Finally, we would really appreciate it if you could find 5 min to fill out our survey, and let us know how the next meeting could be improved, that would be very helpful for the next organising committee and the ancient metagenomics community in general. The deadline for the survey is also October 16 and you will find it here: https://forms.gle/Z6mM9Zt9QXxHvk637
Thanks again for your contribution
Best,
The Organization Team Standards, Precautions & Advances in Ancient Metagenomics 2
--
NEW DATES for the ISBA9 conference in Toulouse NOW June 1st-4th 2021, Don't miss it! International Symposium of Biomolecular Archaeology #ISBA9 https://isba9.sciencesconf.org/
Dr Clio Der Sarkissian CNRS Researcher Team AGES Laboratory AMIS Faculté de Médecine de Purpan 37 allée Jules Guesde 31000 Toulouse```
Hi everyone Just a reminder that you have until this coming Friday October 16 to add your comments to the documents in the shared Google Drive folder: https://drive.google.com/drive/folders/1XPITeI2Sku9WScE85d4suyYXK1OyFgpU?usp=sharing
AND to fill out our survey: https://forms.gle/Z6mM9Zt9QXxHvk637
Thanks again for your contribution!
Best,
The Organization Team Standards, Precautions & Advances in Ancient Metagenomics 2
@channel the comments are now closed, thanks for those who left them!
I'm pleased to announce the slides/notes from our discussions are now all available (and citable 😉 under a CC-BY license with a Zenodo DOI! ) here: https://github.com/SPAAM-community/spaam-event-notes! Be creative how you use them to make new projects, presentations etc!
The feedback from the survey has also been sared with the <#C01BL1HD57T|spaam3-organisers> who will use it to make the next event even better 💪
Did anyone get my email I just sent out about the SPAAM mailing list? My email is crapping out and saying it didn't send anything but it looks like some people may have got it?
Hi to those attending ISBA9! the conference platform will make virtual “lounge rooms” available for random chats and project-oriented discussions. This could be a good opportunity to meet with other members of the SPAAM community (in the limit of 20 participants though). If you’d like to set up a time to meet, let me know we can arrange that and announce the get-together.
*Thread Reply:* YEESSSSSSS ❤️❤️❤️❤️
OK, for those attending ISBA9, what about DAY 3 = THURSDAY JUNE, 3RD for a SPAAM get-together? Thursday looked good as people might be busy chatting with speakers on Day 2-Wednesday after the “Paleo-environment” session and on Day 4-Friday after the “Microbes & Pathogens” session. Now there are 2 options: 1) LUNCH meeting 12:3013:30 2) AFTERWORK meeting 19:0020:00
*Thread Reply:* I am doing an nf-core/eager live demo at 13:00_13:30...
*Thread Reply:* (but could still do it, and I leave halfway through)
*Thread Reply:* @Clio Der Sarkissian I vote meeting at Pyrénées because how cool would it be to have a SPAAM meet up in the mountains!
*Thread Reply:* Not Cassoulet?
*Thread Reply:* OK YES FOOD
*Thread Reply:* (I thought it was a place)
*Thread Reply:* Holy shit that looks amazing
*Thread Reply:* Yes Cassoulet
*Thread Reply:* Pyrenees or Cassoulet? Pyrenees sounds more classy, but I’m not against a bit of fun.
*Thread Reply:* Did you also receive the e-mail to access your platform account?
*Thread Reply:* I thought we were supposed to get access in advance to fix texts so if you got it as well I might be working for nothing…
*Thread Reply:* Yes, I just logged in and that's how I found out about the different palces
*Thread Reply:* Well depends if you're feeling like gaining or burning energy 😆
*Thread Reply:* Bot hare good wit hme
*Thread Reply:* It is indeed a nice platform!
*Thread Reply:* aaargggh, OK, free time for me, not optimal English for us all, but let’s say it doesn’t matter as long as people understand
*Thread Reply:* Huh? Sorry I don't undersand...
*Thread Reply:* (ironicallly)
*Thread Reply:* thanks for explaining (not ironically)
*Thread Reply:* Oh wait, slack didn't tell me about htis message:
> I thought we were supposed to get access in advance to fix texts so if you got it as well I might be working for nothing…
*Thread Reply:* Everything is understandanble to me so far
*Thread Reply:* There is one page not translated but no-one would use it anyway
*Thread Reply:* Profile > A See my participant file
*Thread Reply:* But honestly 🤷
*Thread Reply:* You can work it out anyway
*Thread Reply:* Also noticed:
*Thread Reply:* In the chat box it's still french
*Thread Reply:* Then on hte 'online' page:
*Thread Reply:* But like I said, basically nothing people will ever use probably
*Thread Reply:* (or that they can't understand
*Thread Reply:* @Clio Der Sarkissian somethign that might be a bit more confusing:
On the right - when I started the private chat with Aida
*Thread Reply:* OK, thanks! I’ll let them know
@channel SPAAM2 meet-up at ISBA9??? ☝️
19:00_20:00 CET on Thursday June 3rd on the ISBA9 platform it is then! I’ll let the committee know.
@Clio Der Sarkissian is on TV!
Well done @Miriam Bravo!
Great hair!
Sorry guys, I won't be able to join the meetup tonight due to family reasons. I hope you have a good time!
Really nice talk @Lena G!!!!
Congrats to all of you who presented a talk or a poster at ISBA9 and thanks for keeping discussions alive!
Thanks so much for organising such a fantastic meeting, and hope to see all of you guys in person soon!
Thanks! you’re always welcome in Toulouse!
Fantastic event, really enjoyed, thanks a lot all the speakers and organizers!
Congrats to both @Ele and @Zandra Fagernäs!!!
Thank you for the organizers and to the speakers. It was an informative event.
Today (and every day really) it was mind blowing! Amazing talks, thank you very much!
*Thread Reply:* Starting with the good stuff I see, behind your laptop 💃
*Thread Reply:* That's entirely @aidanva 😂😂😂
*Thread Reply:* Sugars, scissors and a control remote… who can resist…
The cooking class was amazing @Clio Der Sarkissian thank you to ask the organisers!
It was great!!! Thank you @Clio Der Sarkissian !! Such an awesome idea !
Glad you had a good time! we also did!
*Thread Reply:* Absolutely loved it Clio - really well done 👏👏👏
Hi everybody. Thanks for this slack. I am not sure I have to ask in this specific channel concerning some issues I face with nf-core/eager
Hi @pierrespc - genreal questions for SPAAM people is better on #general (this channel is just for discussion about the SPAAM2 workshop from last year). However if you have specific questions about eager, please join the nf-core slack: https://nf-co.re/join and join the #eager channel there 🙂
But good point: I will be *closing* this channel when <#C025AP39256|spaam3-open> happens in September! So please direct general discussions to #general, but of course join <#C025AP39256|spaam3-open> for discussions about the next event 🙂
Hello @channel! The SPAAM2 organizers are pleased to announce that the manuscript draft based on discussions at SPAAM2 last September is now assembled. A major theme we saw throughout the conference and the reviews that were shared with us was that many people struggle to convince people outside the field of the authenticity and relevance of their work. To attempt to ease this issue, we assembled a short opinion piece detailing critical points in ancient metagenomics to assist peer reviewers understand and interpret ancient data.
We invite all SPAAM2 participants to read through and leave comments over the next 2 weeks (last day September 24th). We especially hope the sdeaDNA people will check that we didn't miss anything important as none of us work specifically in that area
Please know that everyone who sent us reviews or review summaries, and everyone who has comments on this draft, will be mentioned by name in the Acknowledgments. If you have questions feel free to message or email us. Here's the link: https://docs.google.com/document/d/17Hfxwsr12_VOsS0cMQn_kfvUnzDQYTidckymrqUqHVc/edit?usp=sharing
*Thread Reply:* No comments, but just wanted to say it was a great read, well don everyone. Table 2 is a great idea!
*Thread Reply:* Same ! It was a really nice recap for someone who just joined the field. Plus, I love figure 2 🤩!
@ivelsko @James Fellows Yates -- have added a few comments to make it more sedaDNA friendly. 🙂
*Thread Reply:* Thank you very much
Great manuscript @ivelsko, @James Fellows Yates and other guys, I read it with lot of interest and added a few comments on computational data analysis challenges, hopefully you will find them useful.
Really enjoyed reading this @ivelsko @James Fellows Yates, well done. I just wanted to write the is awesome everywhere, but felt that would be a little annoying. So I'll say it here instead!
Great paper, I really enjoyed reading it too - it's very clear and easy to read. I've added a few comments based on personal experience working with epidemiologists, but see how you guys feel about them ^^
@Pete Heintzman @Nikolay Oskolkov @Jessica Hider @Katerina Guschanski @Ophélie Lebrasseur I've gone through and made suggestions based on your comments. Please let me know if they work for you (@Clio Der Sarkissian @Alex Hübner @irinavelsko @Åshild (Ash) @Anna F. as well!)
@James Fellows Yates, Just looked through and I'm happy with all the changes/suggestions you've made. Thank you 😊
@irinavelsko, @James Fellows Yates and the rest of SPAAM2 organizers, I also enjoyed reading your article since it is a very much needed information in the field. I made a few irrelevant suggestions 🙂. Thanks for sharing !
Hi! I just had the chance to go through the manuscript, and what a nice read it is! Left a few comments, probably all irrelevant. Great work SPAAM2 organisers!
@channel hive mind, can you give me a list of papers that involve wet lab method development for ancient Metagenomics (either papers specifically addressing it, or studies which included a method dev component)?
The ones from my head are just ones which I've been in the sphere of, so I suspect I'm missing some as wet lab is not my expertise.
Current ones: Warinner 2014, Hagan 2020, Brealey 2020, fagernäs 2020, Farrer 2021
Have I really got everything?
And @Linda Armbrecht’s marine sedaDNAcapture one
@Clio Der Sarkissian did your shell one also include some extraction optiomisation or something 🤔
 }
}




















 }
}











