Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
@Gunnar Neumann has joined the channel
@Maria Lopopolo has joined the channel
question for protocols.io: it seems quite buggy, for example tables cannot be always edited after creation (right click menu to add rows etc.), but my most recent problem/question is: do all authors need to create profiles or can you also add them just as free text? If the latter should be the case I have discovered another bug.
- Yes, some parts of the interface can still be a bit buggy, but they are working on them.
- No, you can add as free text I believe
okay then there’s another bug. it just shows the revolving loading icon instead of the “add” button
Maybe try refreshing the page, or adding a , at the end?
@Katerina Guschanski has joined the channel
Tha'ts normally my solutoin with protocols.io
Still better than anything I've used so far
A lot of manual work, but I’m getting there…
Yeah requires a bit of investment first time
but every subsequent publication: fork, tweak that one step, YOUR HWOLE METHODS SECTION DONE
@Zuzana Hofmanová has joined the channel
@Claudio Ottoni has joined the channel
@Sterling Wright has joined the channel
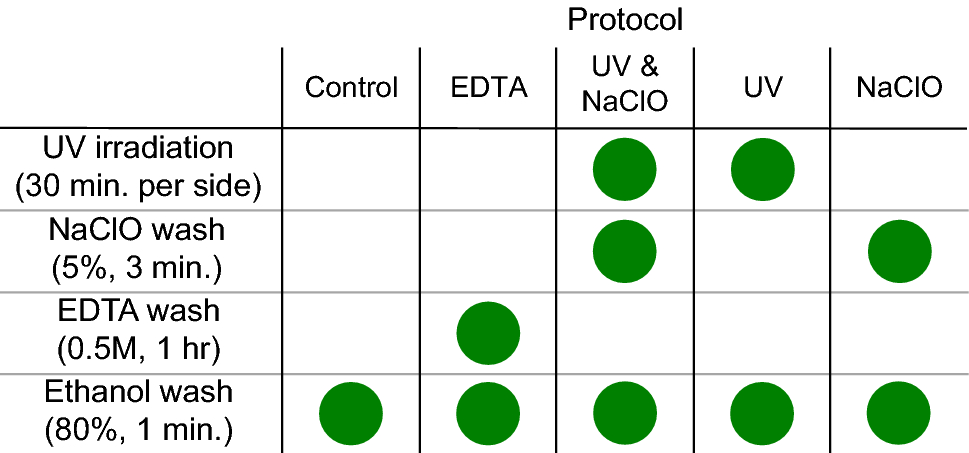
Hi!!! I don’t know if this question falls better here or in <#C02DCKJ54JX|no-stupid-questions> but I guess it would be good for both channels aahha It is a lot that I have this question but I would like your thougths in this.. When you are dealing with dental calculus and you want to study oral microbiome, do you perform a decontamination before the extraction protocol? If yes, what do you use (EDTA wash, bleach and ethanol, some other buffer,..) and why? Why not? Have you tested different decontamination protocols and found statistically significative differences?
Thanks in advance ❤️
*Thread Reply:* We do UV surface decontamination and EDTA wash. Tested it on very few samples and not really systematically, but observed an increase in oral bacs after the treatment compared to before. Results for this are presented here: https://academic.oup.com/mbe/article/37/10/3003/5848415?login=false (check out Fig. S3 in Supp)
*Thread Reply:* Same for us too!
Tested by Andrew and Tina here: Ozga, A. T., Nieves-Colón, M. A., Honap, T. P., Sankaranarayanan, K., Hofman, C. A., Milner, G. R., Lewis, C. M., Jr, Stone, A. C., & Warinner, C. (2016). Successful enrichment and recovery of whole mitochondrial genomes from ancient human dental calculus. American Journal of Physical Anthropology, 160(2), 220–228. https://doi.org/10.1002/ajpa.22960
and our current protocol: https://dx.doi.org/10.17504/protocols.io.bqbmmsk6 (step 9 onwards)
*Thread Reply:* You may be interested in this paper: https://www.nature.com/articles/s41598-021-86100-w
We explored how the commonly used decontamination methods impacted the recovery of oral microbes. It doesn’t seem like to be a huge difference among a couple.
*Thread Reply:* Thank you so much everyone!
☺️
A question for those working with the Santa Cruz Protocol: am I right to assume that this protocol is compatible with the “NEBNext Multiplex Oligos for Illumina (96 Unique Dual Index Primer Pairs)” sets (e.g. NEB E6440S) for indexing?
*Thread Reply:* Correct, the NEB indexing primers are compatible with the SCR.
Hi community, we are thinking of scaling up our wet lab protocols for ancient dental calculus to allow processing more samples at once. We are currently doing everything manually (extractions, library preps, clean-up), sample by sample, not even using a manifold. What are peoples' approaches and kit for multi-sample processing? I think the development has passed me by, so would be happy to hear the best hacks (except that robots are definitely outside of our budget!)