Public Channels
- # 2022-summerschool-introtometagenomics
- # 2023-summerschool-introtometagenomics
- # 2024-acad-aedna-workshop
- # 2024-summerschool-introtometagenomics
- # amdirt-dev
- # analysis-comparison-challenge
- # analysis-reproducibility
- # ancient-metagenomics-labs
- # ancient-microbial-genomics
- # ancient-microbiomes
- # ancientmetagenomedir
- # ancientmetagenomedir-c14-extension
- # authentication-standards
- # benchmark-datasets
- # classifier-committee
- # datasharing
- # de-novo-assembly
- # dir-environmental
- # dir-host-metagenome
- # dir-single-genome
- # eaa-2024-rome
- # early-career-funding-opportunities
- # espaamñol
- # events
- # general
- # it-crowd
- # jobs
- # lab-community
- # lactobacillaceae-spaam4
- # little-book-smiley-plots
- # microbial-genomics
- # minas-environmental
- # minas-metadata-standards
- # minas-microbiome
- # minas-pathogen
- # no-stupid-questions
- # papers
- # random
- # sampling
- # scr-protocol
- # seda-dna
- # spaam-across-the-pond
- # spaam-bingo
- # spaam-blog
- # spaam-ethics
- # spaam-pets
- # spaam-turkish
- # spaam-tv
- # spaam2-open
- # spaam3-open
- # spaam4-open
- # spaam5-open
- # spaam5-organizers
- # spaamfic
- # spaamghetti
- # spaamtisch
- # wetlab_protocols
Private Channels
Direct Messages
Group Direct Messages
@James Fellows Yates has joined the channel
@Clio Der Sarkissian has joined the channel
@Antonio Fernandez-Guerra has joined the channel
@Sterling Wright has joined the channel
For those who often have to generate a taxonomy table of a list of genomes based on the NCBI taxonomy , I just came across this R package which looks VERY nice and might do a lot the heavy work for you. I'm trying it now, so will let you know if it works:
https://github.com/sherrillmix/taxonomizr#preparation
*Thread Reply:* About 60GB in total
*Thread Reply:* Does it update renamed tax info, i.e. replace the taxid with the new taxid if it changed recently?
*Thread Reply:* It looks like that at the moment, you have to re-build the database each time you want to get the latest changes. It just downloads the latest taxdump.tar.gz and then converts that into an SQL datbaase
*Thread Reply:* It took about 20-30 minutes to build on my laptop
*Thread Reply:* OK. I usually use the NCBITaxa() function of the Python module ete3 and it does something similar but it is a pain to just switch over to Python, when I do the rest of the analysis in R. Thanks for pointing that out!
*Thread Reply:* It just finished downloading, so I'll let you know how the other functions work. If it's nice I'll store it centrally on our servers somwhere
*Thread Reply:* Oops, sorry I meant *60* GB
*Thread Reply:* > Does it update renamed tax info, i.e. replace the taxid with the new taxid if it changed recently? It doesn't store old names at least, as well, as I just found out
*Thread Reply:* I am not sure how the Python package does it but sometimes it prints a warning about a new taxid. Nevermind, it barely ever happens.
*Thread Reply:* I used this package very often this year for a metagenomic project. Is really very helpful. For instance, to add the taxonomic path to a krakenUnique result with TaxIDs
@channel If any 'tweeters' here are willing to help, I have a little poll on twitter about what reference genome alignmers you use: https://twitter.com/jafellowsyates/status/1265964157985316864
(note I mean single genome, not metagneomic)
*Thread Reply:* Have you seen the new paper by Marine Poullet and Ludovic Orlando where they compare bwa vs. bowtie2? It would be nice to have both in nf-core EAGER 🙂 (https://www.frontiersin.org/articles/10.3389/fevo.2020.00105/full)
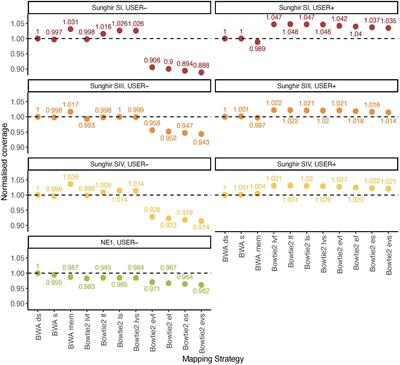
*Thread Reply:* Yes but (copied from another slack workspace):
James Fellows Yates [2:58 PM] I saw that, but found it very confusing James Fellows Yates [2:58 PM] Because it was almost always in the ocntext of methylomes, or at least the focus was always on that James Fellows Yates [2:58 PM] But they also didn't supply any code or commands ot make it easy to work out exactly what they were running James Fellows Yates [2:59 PM] I was excited because I needed something like that to help guide adding bowtie to eager (I'venever used it), but then 🤷 no hepl here James Fellows Yates [3:01 PM] Also check Supplemtnary figure 4 and 5
*Thread Reply:* It's not bad by any means, just it put me off reading in more detail
*Thread Reply:* Would be helpful if they supplied the commands, am reading it in more detail now
Congrats to @Ele on her grant on ancient poop microbiomes!
https://twitter.com/UoYArchaeology/status/1305853141083197441?s=19

*Thread Reply:* Thanks James!!!! Sorry that was slow I haven’t been on slack for a while 😄 Looking forward to getting some tips today and tomorrow 😄
I saw that one of the things that people were interested in was having journal clubs. So I wanted to share that my lab will be hosting an aDNA journal club every two weeks. Our next meeting will be on October 2nd. Feel free to message me if you would like to join.
Hi everyone,
Could I get some advice for human data?
I get the impression that lots of you are interested in the "extra bits" that come from sequencing human samples. Well, I work on sediment and my "extra bits" are human. We want to search for endogenous human sequences. I'm starting with FASTAs containing sequences that MEGAN assigned to Primates.
I've had some success with using PMDtools to filter by damage, but when it comes to aligning to genomes and stuff I have no clue. What's a good place to start?
*Thread Reply:* Not to do any self-PR, but would a pipeline help? https://github.com/nf-core/eager/tree/dev
However that only accepts FASTQs.
And it might be overkill, how many reads have you retrieved?
*Thread Reply:* as James says it depends on the number of reads - you can either do a super quick mapping and profiling against the human reference or do a paleomix or eager run that can do all the steps for you (but might not be necessary if you got say 100 -200 reads)
*Thread Reply:* Also depends what you wanna do after/what you question is, ofc
*Thread Reply:* You also want to check that the regions pulled out are specific to humans and not shared across mammals or eukaryotes. Which dependent on size of the fragments is easier/harder to do.
*Thread Reply:* ☝️ that is also important. I am assuming if you've used MEGAN, that LCA is already applied. But depends what DB you used it.
*Thread Reply:* Ooh, lots to think about! Thanks folks. • I can get FASTQs. • We extracted Primates reads using MEGAN after a quick-and-dirty BLAST against GenBank. • It's going to be something like 10,000-100,000 reads.
*Thread Reply:* 10k-100k sounds like you can go straight into mapping e.g. with eager/paleomix. Or check the BLAST results, and assess breadth of the genome covered (e.g. checking how many of each chromosome it hits, assuming these are the references hit), or how many sub-optimal hits of each read hits non-Homo spapien references
*Thread Reply:* Thank you, I should be able to make a good start with this.
*Thread Reply:* 3' internal barcode that has not been trimmed, perhaps? What library prep and seq. strategy were you using?
*Thread Reply:* ☝️ also from my experience
*Thread Reply:* And @aidanva (sitting next to me) agrees
*Thread Reply:* It is the sample ERR3003613 from Velsko et al., 2019 (https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-019-0717-3). I checked cleaned reads with fastqc. It seems there is no adaptor content and the overal quality looks good. Any other clue? like mapping parameter or coverage unevenness etc ?

*Thread Reply:* or maybe reference quality? since I am using MAG (metagenome asssembled genome), the quality is not as good as isolate genome.
*Thread Reply:* How have you mapped the PE reads, which tool and algorithm (samse/sampe)? did you collapse the PE reads before mapping?
*Thread Reply:* I used bowtie2 (--end-to-end) and I just simply merged PE reads together when mapping. The pipeline worked well before on our internal data.
*Thread Reply:* I am going to give a closer look in mapping procedure.
*Thread Reply:* The only thing that comes to mind for me (if the adapter/barcodes have all been removed) is that if you have the 3' ends of reads being taken into account somehow, and they are not the true end of the reads, you can get a weird mapdamage pattern on the 3' end.
*Thread Reply:* The devil is found ! adaptors in 3' reads were just partially trimmed. Strangely it passed fastqc. There are something wrong with automatic adaptor detecting & trimming. Thanks all you guys! 👍
*Thread Reply:* maybe barcodes, precisely speaking.
*Thread Reply:* First SPAAM collaborative tech-support 💪
*Thread Reply:* tech support
*Thread Reply:* @ivelsko Just in case you haven’t seen this thread, see above
*Thread Reply:* Sorry I missed this. Tip for future support - tag the manuscript first author she can see the thread help you quicker next time
Own-horn tooting: we just released the a new (and 🤞 much more stable) version of nf-core/eager! Note nf-core/eager includes in-parallel metagenomic screening alongside genome mapping and has a bunch of new useful stuff for pathogen genomics.
https://twitter.com/jafellowsyates/status/1318839911357812738
If anyone's lab - or enough from people from SPAAM - is interested in having an introductory workshop, please DM me and we can talk about setting somethign up
*Thread Reply:* Woohoo! Does this mean that all of the parts in dev are now in master?
*Thread Reply:* amazing @James Fellows Yates and team! time to celebrate i hope! 🙌:skintone3:
*Thread Reply:* Need to submit the paper first 😆
*Thread Reply:* pfff - super minor - take every step as a win !!
@channel just a small reminder, if anyone is interested in having an introductory workshop for nf-core/eager, please let me know via DM. I have 11 people who would be interested. If I reach 15 I will start organising
*Thread Reply:* Hey James, if do organize it, the PhD student of the lab will probably like to do it
*Thread Reply:* Ok, in principle 2 more people (if I include yourself with the PhD student ;)) then happy to run it
No, you do not know what is 'relevant' for other people 😠 /rant
*Thread Reply:* Sedi-DNA Denosivan stuff from Qiaomei
*Thread Reply:* Yeah just mtDNA sequences loaded
*Thread Reply:* It’s not really reproducible then
*Thread Reply:* Although I guess it's from China might also be some bureaucratic issues
Bioinformatics folks in particular, I was chatting to a friend who helps run the Society of Research Software Engineering. It's an organisation for software developers in academia. Might be another helpful community for some of us 🙂
*Thread Reply:* should we invite them to the slack channel ? 🤔
*Thread Reply:* If we can find contact details, go for it!
@channel Any suggestions for good independent reviewers in museum genomics? or aDNA for museums? Asking for a funding panel 🙂
*Thread Reply:* @Katerina Guschanski and @Rita M Austin might know some, I think
*Thread Reply:* Was just thinking. I guess "good" and "independent" depends a lot on where you stand. I'd suggest Tom Gilbert, Andrew Foote, Love Dalén. If plants Hernan Burbano.
*Thread Reply:* Iirc Beth Shapiro has dabbled in it (or at least worked with people who has)
*Thread Reply:* And she had the passenger pigeon thing
*Thread Reply:* Independent in the sense that they are an external scientific reviewer
*Thread Reply:* Selina Brace and Ian Barnes at the NHM London 🙂
*Thread Reply:* I can suggest Logan Kistler, Peter Heintzman, Inger Alsos, and Audrey Lin.
Hey all! The Penn State Ancient Biomolecules Research Group is running a survey designed to understand how dental calculus is sampled and what metadata is collected. If you have time, please fill out this quick survey https://pennstate.qualtrics.com/jfe/form/SV_8pniw3vF7565UxL and forward it to anyone you know who does dental calculus work! Thanks you!!!!
*Thread Reply:* You could also post to spaam-community@listserv.dfn.de and there is also an ancient DNA mailing list in the UK you could also try 🙂 https://www.jiscmail.ac.uk/cgi-bin/webadmin?A0=ANCIENT-DNA
If there is anyone using paperpile, and have a 'personal' account (or want to switch from an institutional-locked one to personal to take your account with you), I have a 'forever' discount code for 20% that you could use if you want.
(Context: IT at our institute forgot to renew licenses, and a lot of people considered switching to personal ones as it's only ~€30 a year, and when I said we had like ~10 people considering it, they offered me a 'ambassador' code because I've been with them for >5 years).
I'm not going to push/sell this, just wanted to offer it to people outside our institute (therefore feel free to pass on the message to any of your colleagues)!
*Thread Reply:* @James Fellows Yates I’d like the code! Been thinking of switching to paperpile and abandoning endNote
Hi all, just a reminder to please fill out this SUPER QUICK survey about dental calculus sampling if you work with this material! https://pennstate.qualtrics.com/jfe/form/SV_8pniw3vF7565UxL We really appreciate it 😄
@channel if any of you are presenting at ISBA9, @Betsy Nelson had a great idea on Twitter this morning (or evening, or whatever space time disruption we live in):
https://twitter.com/eanelson42/status/1395599481215897603?s=19
(It was also just endorsed by Ludovic as well ;))
*Thread Reply:* Brilliant! 🤣
*Thread Reply:* Oh you guys included your faces!? I’m just a disembodied voice…
*Thread Reply:* Oh I gave up with video in the end, it was taking too long 😆
*Thread Reply:* PLease do share!!!
Had a lot of fun watching those at the meeting of the ISBA organisation committee this morning! Thanks for the nice break!
*Thread Reply:* 😅😅😅 we need moar!!!
*Thread Reply:* #ISBA9VideoBlooper now official (look at your emails)
In addition to the blooper reel, if anyone is tired of the same pop-gen talk over and over again just on a different region of the world, I've modified a little game I did for a different event for ISBA9:
https://twitter.com/jfy133/status/1399331712409247745

And if you people can suggest (minimum) 25 cards for Ancient Metagenomics, feel free to send them me and I'll make an ancientmetagenomics one!
e.g. "_Yersinia pestis is the aetological agent of plague" in the first 30s of a pathogen talk
Some genome reconstruction with coverage plot
And of course damage patterns 🙂
Thanks to @Zandra Fagernäs for initially retweeting 😂😂😂
https://twitter.com/JMA_Stough/status/1399108474655674373?s=19

Random R / tidyverse tip for the day of something I only just learnt (but might be useful to others if they haven't already found it):
If you ever want to find the row after something you filter for in a table (e.g. debugging which row a function fails on), you can use lag()
```> hostasssamacc %>% filter(lag(projectname == "Ottoni2019"))
A tibble: 10 x 69
projectname publicationyear publicationdoi sitename latitude longitude geolocname samplename samplehost sampleage sampleage_doi
<chr> <dbl> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB05 Papio anubis 100 10.1038/s41598…
2 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB05 Papio anubis 100 10.1038/s41598…
3 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB06 Papio anubis 100 10.1038/s41598…
4 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB06 Papio anubis 100 10.1038/s41598…
5 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB08 Papio hamad… 100 10.1038/s41598…
6 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB09 Papio hamad… 100 10.1038/s41598…
7 Ottoni2019 2019 10.1038/s41598-0… Gabbanat al-… 25.7 32.6 Egypt BB11 Papio sp. 100 10.1038/s41598…
8 Ottoni2019 2019 10.1038/s41598-0… Zoological S… 40.0 -75.2 USA ANSP11833 Papio hamad… 100 10.1038/s41598…
9 Ottoni2019 2019 10.1038/s41598-0… Zoological S… 40.0 -75.2 USA ANSP3271 Papio hamad… 100 10.1038/s41598…
10 Zhou2018 2018 10.1016/j.cub.20… St. Olav's C… 63.4 10.4 Norway SK152 Homo sapiens 800 10.1016/j.cub.…```
You can see I searched for Ottoni, but also got the next row which was Zhou
Anyone have access to this publication? Ennever J Steigenberger O.P. & Radike, A.W. (1961) The calculus surface index method for scoring clinical calculus studies. Journal of periodontology
Congrats to Dra. @Miriam Bravo!!
https://twitter.com/paleogenomics/status/1422731114410659844?s=19

Can anyone recommend something that nicely explains the lambda DNA fragmentation metric and how to interpret it? Maybe a nice blog post or something?
*Thread Reply:* Good question!
*Thread Reply:* I don't htink I've any 🤔
*Thread Reply:* Maybe we could ask someone to write one for us 😬
*Thread Reply:* Thanks for the quick reply James! Ran the scripts, got the numbers just trying figure out what they mean 🤦♀️
*Thread Reply:* That's not so desciprtive is it 😆
*Thread Reply:* Guys, are you talking about one of the metrics returned by mapDamage?
*Thread Reply:* That's what I thought what @Ele was referring to, at least
*Thread Reply:* But it's been used elsewhere
*Thread Reply:* E.g. thermal age I believe (might be wrong there though)
*Thread Reply:* Ok, if @Ele was referring to the lambda parameter from mapDamage I could try to give some interpretation but if she meant something else then I probably unfortunately can't help
*Thread Reply:* I would be curious regardles!
*Thread Reply:* Reason number 7892542 of confusion! I actually meant the thermal age one (specifically used in Kistler et al 17:https://academic.oup.com/nar/article/45/11/6310/3806656 https://academic.oup.com/nar/article/45/11/6310/3806656 basically looks at rate of fragmentation based on a curve and spits a number. I think a bigger number = a steeper curve = more rapid fragmentation. But it’s that “a bigger number” that’s causing me trouble… bigger than..?
*Thread Reply:* But yes @Nikolay Oskolkov defo interested in hearing more about mapdamage lambda because there’s another metric I’m confused by. I’d take a theta explanation too hahaha
*Thread Reply:* Ok, after having had a quick look at the Kistler et al., I believe the lambda @Ele was asking about is nothing more than just a typical DNA fragment length, and this is very different from the lambda (and theta) reported by mapDamage.
In Kistler et al., they model fragment length distribution via exponential distribution as ~exp(- x / lambda), where x is a length of a particular fragment, and lambda is a "typical" / mean / median fragment length. As simple as that.
In contrast, the lambda parameter estimated within the mapDamage framework is a sort of probability that a query cytosine (i.e. the one that potentially can be deaminated) is within an overhang region (if I understand correctly) on a DNA fragment. This lambda probability is related to the length of an overhang, i.e. the longer is an overhang the greater is the probability that a cytosine on the overhang is deaminated. There is another parameter "nu", that is a nick frequency and therefore a probability that the query cytosine falls onto a nick region.
The theta parameter is actually a posterior probability that the query cytosine is mutated not due to damage. In the mapDamage paper they give a joint posterior probability of a position-specific mutation (for any nucleotide) as a product of probability of true biological / germline mutations (theta, not due to damage) and probability of damage mutations (Pdam). Theta is computed exactly as any other Markov model of nucleotide substitutions (JC69, K80, F81, HKY85 etc.), i.e. as a 4x4 matrix of probabilities of a nucleotide to mutate into A, C, T or G. So theta has nothing to do with damage, while Pdam is computed as a 4x4 matrix of probabilities of C to mutate into T (expressed via lambda and nu) and G to mutate to A.
Actually, the strength of mapDamage is the statistical framework, however many of us use it just for plotting the deamination pattern and ignore the tones of useful metrics (unless you are a fan of Bayesian way of analysis). Many other tools (like PMDtools) can plot the deamination pattern but they can't give you all those useful probability estimates
*Thread Reply:* @Nikolay Oskolkov wanna make a blogpost 😬 ?
*Thread Reply:* The tragedy of academic science is that our articles are too complex 🙂 (even if you are not a newbie), and reading a scientific paper is like decoding an enigma, takes hours (personally for me). Just think, one needs to write blog posts explaining what was written in the articles which I think is a sort of shame. Why not to directly write understandable articles? 🙂
*Thread Reply:* I completely agree!
*Thread Reply:* (was suggesting if you wanted to write a short one as you are very good at doing them : ))
*Thread Reply:* (but no worries if too much work)
*Thread Reply:* (but what yu've written above is a very good summary I tink)
*Thread Reply:* yes, if there an interest, I could try to expand the summary above. Then it should probably be a practical example where one could start from a substitution-rate matrix and infer the pairwise nucleotide substitution probabilities by matching the model to the data via maximum likelihood. There are many plans but too little time 😞
*Thread Reply:* Well if you ever found a moment 😉
*Thread Reply:* Thank you so much for the explanation @Nikolay Oskolkov that’s really helpful. I’ll go through the paper again and try to see if I can piece all the bits together. You’re a star!
*Thread Reply:* What if SPAAM starts a blog exactly for this purpose? It could post short informative answers to questions as they come up?
*Thread Reply:* Good idea!
@Nikolay Oskolkov would you be interested in inaugurating such a thing?
*Thread Reply:* Could also come up with a way of archiving it on Zenodo so one can get DOIs and cite it as well 🤔
*Thread Reply:* @James Fellows Yates and @Christina Warinner I would be interested, yes. How and where would you suggest to arrange such a blog?
*Thread Reply:* I'm open to ideas, but my initial thought would be I set up a github repository, and we host in the form of markdown files
*Thread Reply:* Because then we can have it on the spaam website, but also do the archiving
*Thread Reply:* (and then easier to share access across multiple people)
*Thread Reply:* Maybe I ask in #general, is this would be ok with you?
*Thread Reply:* Sure @James Fellows Yates !
Does anyone have krakenuniq database building files? I'm working on building a database and am encountering issues, and it would be useful to have sample files
*Thread Reply:* Abby please check what files are downloaded here:
```## Download the taxonomy krakenuniq-download --db DBDIR taxonomy
All microbial (including eukaryotes) sequences in the NCBI nt database
krakenuniq-download --db DBDIR --dust microbial-nt``` It should be not only the fasta-file but also taxonomy mapped to each of the reference genomes
Would anyone be interested in setting up a 'music league' for SPAAM?
Each week there is a theme, you submit songs from Spotify matching the theme somehow, once everyone submits you get a playlist and can vote for your favourites (whoever submitted the most voted song gets to choose the next theme)
*Thread Reply:* please add me : eric.capo@hotmail.fr by the way, here is a playlist I am listening a lot when coding and writing 🙂 https://www.youtube.com/watch?v=G-I5GN5XCKs
*Thread Reply:* oh yes! that would be fun!!
*Thread Reply:* should we rebrand
*Thread Reply:* I was wondering the same 😂
Just came across this while deleting my google scholar notifications...: Are you a timetraveller @Miriam Bravo!!?!?
(from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9791188/)

Don't blindly download NT/Refsrq people!
RT @voorloopnul In today's date, ~1.7 million bacterial genomes are being hosted in NCBI.
Salmonella Enterica and Escherichia Coli represent around 40% of those genomes.
Considering only the species that make 1% or more , we have 12 bacteria summing to ~64% of all bacterial genomes.
https://mstdn.science/@EvolvedBiofilm/109657985446766965
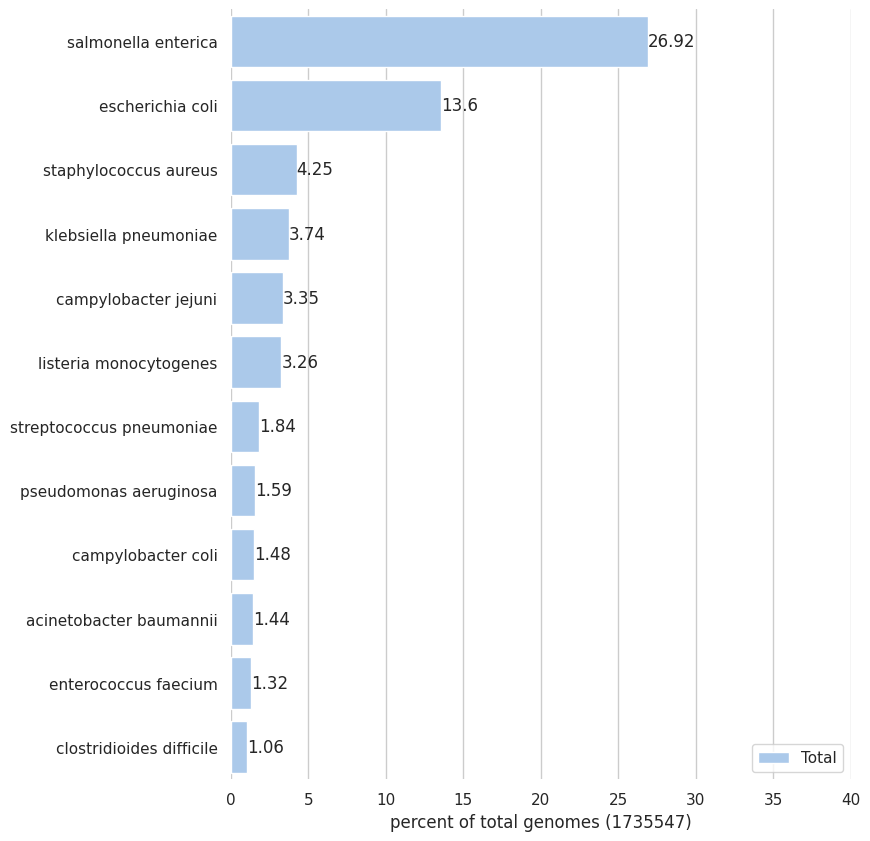
*Thread Reply:* Very interesting! I wonder why Salmonella Enterica? 🤔 Also, among overrepresented species there are so many pathogenic ones 😞
*Thread Reply:* Pathogenic because of clinical interest/epidemiology
*Thread Reply:* S. Enterica in particular I'm not sure why, maybe easy to isolate/culture and common pathogen? But I'm not a microbiologist... @irinavelsko would you happen to know?
*Thread Reply:* Both are model organisms with historical precedence for molecular studies. They’re easy to grow and easy to genetically manipulate. Salmonella is probably particularly a problem for food-borne diseases so there’s clinical and epidemiological interest from that angle
*Thread Reply:* I think also S enterica has thousands of serovars so it makes sense that many would be sequenced
Made by Kathrin Nägele
🚨Hello @channel🚨 📣Check out the FIRST SPAAM :spaam: NEWSLETTER!! 📣 :headbangingparrot: We will have the next one in three months! :spaam::catjam::awwyeah:
The good kind of SPAAM in my Thunderbird inbox 😉
Today at EAA2023 we had a small unexpected SPAAM gathering and here a picture of @Zoé Pochon presenting her work!! 😍❤️
*Thread Reply:* Do you have a group photo?!
*Thread Reply:* We should have taken one, but didn't think of it 😅
*Thread Reply:* Oh well! Maybe we should make it a rule 😬
*Thread Reply:* Also maybe make more stickers and distribute packs around the world to people to give out at ninja-SPAAM meetings!
*Thread Reply:* We will make it a rule for sure 😍 And we should for sure have more stickers!! If you have the file I can print some more!! :spaamtisch:
*Thread Reply:* All the logos are in the website!
*Thread Reply:* But I can send you a link to the company I've done all my sticks for, with a reording link, one sec
*Thread Reply:* Ah that I don't have anymore
*Thread Reply:* But this is the company: https://stickerapp.de/ (they have websites in multiple EU countries)
*Thread Reply:* All logo variants are there 👆
*Thread Reply:* Great!! 😍😍
Gonna start bringing some around
*Thread Reply:* But that's examples of others I've done
*Thread Reply:* I also have made some stickers for AMDirT 🙂
*Thread Reply:* spotted 😰
Hello! Is there a SPAAM template for slides that you normally use?
*Thread Reply:* Open this so it's registered in your drive
*Thread Reply:* Then go to your slides
*Thread Reply:* And then theme then import
*Thread Reply:* Yay! It worked! Thanks James ❤️

*Thread Reply:* Hey @Martina Farese, Have a look at <#C05TBKJ5HU1|eaa-2024-rome>;)
spaam-skaating? Feat @irinavelsko and @Ophélie Lebrasseur (or @Ophélie Lebrasseur)
Why we should maybe move from slack ..
amapanda@en.osm.town - Ouch. A large OpenStreetMap group has been using a proprietary chat platform as a community space for ~10 yrs. Now they gotta pay a $80k/yr (or $10k??) for usage. 🤯😱😢
Slack (now Salesforce) now wants to charge @OpenStreetMapUS for all ~6k users on their server. 😢😢 Ouch.
This sort of bait & switch is why open, community owned platforms (like this!) are vital!
read more on the slack (while you still can??!): https://osmus.slack.com/archives/C029HV951/p1705438543546349
OpenStreetMap #OSM #FreeSoftware #Cassandra
(From mastodon)
*Thread Reply:* It's time to enter the Matrix! 😆
*Thread Reply:* But seriously, chat.archaeo.social may be the way to go
*Thread Reply:* (just an experiment though ATM)
 }
}


 }
}
 }
}

 }
}

 }
}
 }
}


 }
}











